[English] 日本語
 Yorodumi
Yorodumi- EMDB-30450: Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30450 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
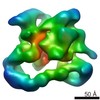
| Title | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NADase / ETI / HR / PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationeffector-mediated perturbation of host process by symbiont / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / response to other organism / NAD+ nucleosidase activity, cyclic ADP-ribose generating / defense response / ADP binding / host cell cytoplasm / host cell nucleus / signal transduction / extracellular region / ATP binding Similarity search - Function | |||||||||
| Biological species |   Hyaloperonospora arabidopsidis (strain Emoy2) (eukaryote) Hyaloperonospora arabidopsidis (strain Emoy2) (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Ma SC / Lapin D | |||||||||
| Funding support |  China, China,  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Authors: Shoucai Ma / Dmitry Lapin / Li Liu / Yue Sun / Wen Song / Xiaoxiao Zhang / Elke Logemann / Dongli Yu / Jia Wang / Jan Jirschitzka / Zhifu Han / Paul Schulze-Lefert / Jane E Parker / Jijie Chai /   Abstract: Direct or indirect recognition of pathogen-derived effectors by plant nucleotide-binding leucine-rich repeat (LRR) receptors (NLRs) initiates innate immune responses. The effector ATR1 activates the ...Direct or indirect recognition of pathogen-derived effectors by plant nucleotide-binding leucine-rich repeat (LRR) receptors (NLRs) initiates innate immune responses. The effector ATR1 activates the N-terminal Toll-interleukin-1 receptor (TIR) domain of NLR RPP1. We report a cryo-electron microscopy structure of RPP1 bound by ATR1. The structure reveals a C-terminal jelly roll/Ig-like domain (C-JID) for specific ATR1 recognition. Biochemical and functional analyses show that ATR1 binds to the C-JID and the LRRs to induce an RPP1 tetrameric assembly required for nicotinamide adenine dinucleotide hydrolase (NADase) activity. RPP1 tetramerization creates two potential active sites, each formed by an asymmetric TIR homodimer. Our data define the mechanism of direct effector recognition by a plant NLR leading to formation of a signaling-active holoenzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30450.map.gz emd_30450.map.gz | 6.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30450-v30.xml emd-30450-v30.xml emd-30450.xml emd-30450.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30450.png emd_30450.png | 54.6 KB | ||
| Filedesc metadata |  emd-30450.cif.gz emd-30450.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30450 http://ftp.pdbj.org/pub/emdb/structures/EMD-30450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30450 | HTTPS FTP |
-Related structure data
| Related structure data |  7crcMC  7crbC  7dfvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30450.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30450.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0979 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tetrameric complex of RPP1 and ATR1
| Entire | Name: Tetrameric complex of RPP1 and ATR1 |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric complex of RPP1 and ATR1
| Supramolecule | Name: Tetrameric complex of RPP1 and ATR1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: RPP1
| Supramolecule | Name: RPP1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: ATR1
| Supramolecule | Name: ATR1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Hyaloperonospora arabidopsidis (strain Emoy2) (eukaryote) Hyaloperonospora arabidopsidis (strain Emoy2) (eukaryote) |
-Macromolecule #1: NAD+ hydrolase (NADase)
| Macromolecule | Name: NAD+ hydrolase (NADase) / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 139.009781 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSAMSLGCS KRKATNQDVD SESRKRRKIC STNDAENCRF IQDESSWKHP WSLCANSVVN DTKDTKSSAL SLPSPPTSVS RIWKHQVFP SFHGADVRKT ILSHILESFR RKGIDPFIDN NIERSKSIGH ELKEAIKGSK IAIVLLSKNY ASSSWCLDEL A EIMKCREL ...String: MGSAMSLGCS KRKATNQDVD SESRKRRKIC STNDAENCRF IQDESSWKHP WSLCANSVVN DTKDTKSSAL SLPSPPTSVS RIWKHQVFP SFHGADVRKT ILSHILESFR RKGIDPFIDN NIERSKSIGH ELKEAIKGSK IAIVLLSKNY ASSSWCLDEL A EIMKCREL LGQIVMTIFY EVDPTDIKKQ TGEFGKAFTK TCKGKTKEYV ERWRKALEDV ATIAGYHSHK WRNEADMIEK IA TDVSNML NSFKPSRDFN GLVGMRAHMD MLEQLLRLVL DEVRMIGIWG PPGIGKTTIA RFLFNQVSDR FQLSAIMVNI KGC YPRPCF DEYSAQLQLQ NQMLSQMINH KDIMISHLGV AQERLRDKKV FLVLDEVDQL GQLDALAKET RWFGPGSRII ITTE DLGVL KAHGINHVYK VGYPSNDEAF QIFCMNAFGQ KQPHEGFDEI AREVMALAGE LPLGLKVLGS ALRGKSKPEW ERTLP RLKT SLDGKIGSII QFSYDALCDE DKYLFLYIAC LFNKESTTKV EGLLGKFLDV RQGLHILAQK SLISIEDGNI YMHTLL EQF GRETSRKQFI HHGYTKHQLL VGERDICEVL NDDTIDSRRF IGINLDLYKN VEELNISEKA LERIHDFQFV RINGKNH AL HERLQGLIYQ SPQIRSLHWK CYQNICLPST FNSEFLVELD MSFSKLQKLW EGTKQLRNLK WMDLSYSSYL KELPNLST A TNLEELKLRN CSSLVELPSS IEKLTSLQIL DLHRCSSLVE LPSFGNATKL EILNLENCSS LVKLPPSINA NNLQELSLT NCSRVVELPA IENATNLWKL NLLNCSSLIE LPLSIGTATN LKHLDFRGCS SLVKLPSSIG DMTNLEVFYL SNCSNLVELP SSIGNLRKL TLLLMRGCSK LETLPTNINL KSLHTLNLID CSRLKSFPEI STHIKYLRLI GTAIKEVPLS IMSWSPLAHF Q ISYFESLK EFPHALDIIT ELQLSKDIQE VPPWVKRMSR LRALRLNNCN NLVSLPQLPD SLAYLYADNC KSLERLDCCF NN PEIRLYF PKCFKLNQEA RDLIMHTSTR NFAMLPGTQV PACFNHRATS GDSLKIKLKE SPLPTTLTFK ACIMLVNEEM SYD LKSMSV DIVIRDEQND LKVQCTPSYH QCTEIYVLTE HIYTFELEVE EVTSTELVFE FTSVNESICK IGECGILQRE TRSL RRSSS PDLSPESSRV SSCDHC UniProtKB: ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase |
-Macromolecule #2: Avirulence protein ATR1
| Macromolecule | Name: Avirulence protein ATR1 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hyaloperonospora arabidopsidis (strain Emoy2) (eukaryote) Hyaloperonospora arabidopsidis (strain Emoy2) (eukaryote) |
| Molecular weight | Theoretical: 35.149426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRVCYFVLVP SVALAVIATE SSETSGTIVH VFPLRDVADH RNDALINRAL RAQTALDDDE ERWPFGPSAV EALIETIDRH GRVSLNDEA KMKKVVRTWK KLIERDDLIG EIGKHYFEAP GPLHDTYDEA LATRLVTTYS DRGVARAILH TRPSDPLSKK A GQAHRLEE ...String: MRVCYFVLVP SVALAVIATE SSETSGTIVH VFPLRDVADH RNDALINRAL RAQTALDDDE ERWPFGPSAV EALIETIDRH GRVSLNDEA KMKKVVRTWK KLIERDDLIG EIGKHYFEAP GPLHDTYDEA LATRLVTTYS DRGVARAILH TRPSDPLSKK A GQAHRLEE AVASLWKGRG YTSDNVVSSI ATGHDVDFFA PTAFTFLVKC VESEDDANNA IFEYFGSNPS RYFSAVLHAM EK PDADSRV LESSKKWMFQ CYAQKQFPTP VFERTLAAYQ SEDYAIRGAR NHYEKLSLSQ IEELVEEYSR IYSV UniProtKB: Avirulence protein ATR1 |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 23.542 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)





















