[English] 日本語
 Yorodumi
Yorodumi- EMDB-28557: Structure of Xenopus cholinephosphotransferase1 in complex with C... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Xenopus cholinephosphotransferase1 in complex with CDP-choline | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | CDP-APs / phospholipid synthesis / TRANSFERASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationdiacylglycerol cholinephosphotransferase / diacylglycerol cholinephosphotransferase activity / Golgi membrane / endoplasmic reticulum membrane / Golgi apparatus / metal ion binding Similarity search - Function | ||||||||||||
| Biological species | |||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Wang L / Zhou M | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of a eukaryotic cholinephosphotransferase-1 reveals mechanisms of substrate recognition and catalysis. Authors: Lie Wang / Ming Zhou /  Abstract: Phosphatidylcholine (PC) is the most abundant phospholipid in eukaryotic cell membranes. In eukaryotes, two highly homologous enzymes, cholinephosphotransferase-1 (CHPT1) and choline/ethanolamine ...Phosphatidylcholine (PC) is the most abundant phospholipid in eukaryotic cell membranes. In eukaryotes, two highly homologous enzymes, cholinephosphotransferase-1 (CHPT1) and choline/ethanolamine phosphotransferase-1 (CEPT1) catalyze the final step of de novo PC synthesis. CHPT1/CEPT1 joins two substrates, cytidine diphosphate-choline (CDP-choline) and diacylglycerol (DAG), to produce PC, and Mg is required for the reaction. However, mechanisms of substrate recognition and catalysis remain unresolved. Here we report structures of a CHPT1 from Xenopus laevis (xlCHPT1) determined by cryo-electron microscopy to an overall resolution of ~3.2 Å. xlCHPT1 forms a homodimer, and each protomer has 10 transmembrane helices (TMs). The first 6 TMs carve out a cone-shaped enclosure in the membrane in which the catalysis occurs. The enclosure opens to the cytosolic side, where a CDP-choline and two Mg are coordinated. The structures identify a catalytic site unique to eukaryotic CHPT1/CEPT1 and suggest an entryway for DAG. The structures also reveal an internal pseudo two-fold symmetry between TM3-6 and TM7-10, and suggest that CHPT1/CEPT1 may have evolved from their distant prokaryotic ancestors through gene duplication. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28557.map.gz emd_28557.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28557-v30.xml emd-28557-v30.xml emd-28557.xml emd-28557.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28557.png emd_28557.png | 35.9 KB | ||
| Filedesc metadata |  emd-28557.cif.gz emd-28557.cif.gz | 6.5 KB | ||
| Others |  emd_28557_half_map_1.map.gz emd_28557_half_map_1.map.gz emd_28557_half_map_2.map.gz emd_28557_half_map_2.map.gz | 27.7 MB 27.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28557 http://ftp.pdbj.org/pub/emdb/structures/EMD-28557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28557 | HTTPS FTP |
-Related structure data
| Related structure data |  8erpMC  8eroC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28557.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28557.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
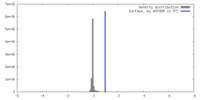
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28557_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
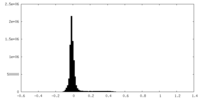
| Density Histograms |
-Half map: #1
| File | emd_28557_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : homodimer of choline phosphotransferase 1
| Entire | Name: homodimer of choline phosphotransferase 1 |
|---|---|
| Components |
|
-Supramolecule #1: homodimer of choline phosphotransferase 1
| Supramolecule | Name: homodimer of choline phosphotransferase 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: Cholinephosphotransferase 1
| Macromolecule | Name: Cholinephosphotransferase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: diacylglycerol cholinephosphotransferase |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 44.54223 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGLAEGLAAR MAPHLYIQEP LSAQQLKKLE EHKYSASGRS LVEPPMQVYW NWLVEKVPLW LAPNTITMVG LLLNVLSTLI LVCYCPTAT EGAPFWTYLL CAIGLFVYQS LDAIDGKQAR RTNSSSPLGE MFDHGCDSIS IVFVNLGTIA AVRLGTLPGW M FYCCFVGM ...String: MGLAEGLAAR MAPHLYIQEP LSAQQLKKLE EHKYSASGRS LVEPPMQVYW NWLVEKVPLW LAPNTITMVG LLLNVLSTLI LVCYCPTAT EGAPFWTYLL CAIGLFVYQS LDAIDGKQAR RTNSSSPLGE MFDHGCDSIS IVFVNLGTIA AVRLGTLPGW M FYCCFVGM FMFYCAQWQT YVCGTLKFGI IDVTELQISV TVMFLMTAVC GPELWDYEIP FTGLPMKTIP LLGIIGGTVY SC SNYFRVI LSGGVGKNGS TVAGTSVLSP GLHIGLVLLL ALMIYKKSTT NLFLQNPCLY TLAFGFVSAK ITIKLVIAHM TKS EISLQD TAFIGPGLLF FNQYFNSFID EYIVLWIAMV ISFADLLRYC ISVCLQIATH LRISVFRISS NQAAEQVQTQ KQKL TD UniProtKB: Cholinephosphotransferase 1 |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 3 / Number of copies: 20 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Macromolecule #4: [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM
| Macromolecule | Name: [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM type: ligand / ID: 4 / Number of copies: 2 / Formula: CDC |
|---|---|
| Molecular weight | Theoretical: 488.324 Da |
| Chemical component information | 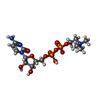 ChemComp-CDC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 8.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)