[English] 日本語
 Yorodumi
Yorodumi- EMDB-28192: Cryo-EM structure of a potent anti-malarial antibody L9 in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a potent anti-malarial antibody L9 in complex with Plasmodium falciparum circumsporozoite protein (PfCSP)(dominant class) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Anti-Malarial / PfCSP / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell surface binding / symbiont entry into host / entry into host cell by a symbiont-containing vacuole / heparan sulfate proteoglycan binding / side of membrane / cell surface / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
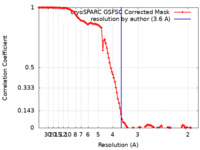
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Tripathi P / Kwong PD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Cryo-EM structures of anti-malarial antibody L9 with circumsporozoite protein reveal trimeric L9 association and complete 27-residue epitope. Authors: Prabhanshu Tripathi / Michael F Bender / Haotian Lei / Lais Da Silva Pereira / Chen-Hsiang Shen / Brian Bonilla / Marlon Dillon / Li Ou / Marie Pancera / Lawrence T Wang / Baoshan Zhang / ...Authors: Prabhanshu Tripathi / Michael F Bender / Haotian Lei / Lais Da Silva Pereira / Chen-Hsiang Shen / Brian Bonilla / Marlon Dillon / Li Ou / Marie Pancera / Lawrence T Wang / Baoshan Zhang / Facundo D Batista / Azza H Idris / Robert A Seder / Peter D Kwong /  Abstract: Monoclonal antibody L9 recognizes the Plasmodium falciparum circumsporozoite protein (PfCSP) and is highly protective following controlled human malaria challenge. To gain insight into its function, ...Monoclonal antibody L9 recognizes the Plasmodium falciparum circumsporozoite protein (PfCSP) and is highly protective following controlled human malaria challenge. To gain insight into its function, we determined cryoelectron microscopy (cryo-EM) structures of L9 in complex with full-length PfCSP and assessed how this recognition influenced protection by wild-type and mutant L9s. Cryo-EM reconstructions at 3.6- and 3.7-Å resolution revealed L9 to recognize PfCSP as an atypical trimer. Each of the three L9s in the trimer directly recognized an Asn-Pro-Asn-Val (NPNV) tetrapeptide on PfCSP and interacted homotypically to facilitate L9-trimer assembly. We analyzed peptides containing different repeat tetrapeptides for binding to wild-type and mutant L9s to delineate epitope and homotypic components of L9 recognition; we found both components necessary for potent malaria protection. Last, we found the 27-residue stretch recognized by L9 to be highly conserved in P. falciparum isolates, suggesting the newly revealed complete L9 epitope to be an attractive vaccine target. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28192.map.gz emd_28192.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28192-v30.xml emd-28192-v30.xml emd-28192.xml emd-28192.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28192_fsc.xml emd_28192_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28192.png emd_28192.png | 36.8 KB | ||
| Filedesc metadata |  emd-28192.cif.gz emd-28192.cif.gz | 5.8 KB | ||
| Others |  emd_28192_half_map_1.map.gz emd_28192_half_map_1.map.gz emd_28192_half_map_2.map.gz emd_28192_half_map_2.map.gz | 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28192 http://ftp.pdbj.org/pub/emdb/structures/EMD-28192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28192 | HTTPS FTP |
-Validation report
| Summary document |  emd_28192_validation.pdf.gz emd_28192_validation.pdf.gz | 847.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28192_full_validation.pdf.gz emd_28192_full_validation.pdf.gz | 846.6 KB | Display | |
| Data in XML |  emd_28192_validation.xml.gz emd_28192_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_28192_validation.cif.gz emd_28192_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28192 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28192 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28192 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28192 | HTTPS FTP |
-Related structure data
| Related structure data |  8ek1MC  8ekaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28192.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28192.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28192_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
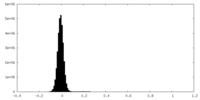
| Density Histograms |
-Half map: #1
| File | emd_28192_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
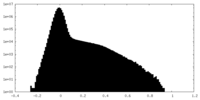
| Density Histograms |
- Sample components
Sample components
-Entire : PfCSP in complex with antibody L9
| Entire | Name: PfCSP in complex with antibody L9 |
|---|---|
| Components |
|
-Supramolecule #1: PfCSP in complex with antibody L9
| Supramolecule | Name: PfCSP in complex with antibody L9 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: L9 Fab heavy chain
| Macromolecule | Name: L9 Fab heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.475436 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVKLVESGGG VVQPGRSLRL SCEASGFIFS TYGMHWVRQA PGKGLEWVAV IWFDGSNIYY ADSVKGRFTI SRDNSKNTVF MQMDSLRAE DTAVYYCHRN FYDGSGPFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS ...String: QVKLVESGGG VVQPGRSLRL SCEASGFIFS TYGMHWVRQA PGKGLEWVAV IWFDGSNIYY ADSVKGRFTI SRDNSKNTVF MQMDSLRAE DTAVYYCHRN FYDGSGPFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCDKTH |
-Macromolecule #2: L9 Fab light chain
| Macromolecule | Name: L9 Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.597254 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPST LSASVGDRVT ITCRASQFIS RWLAWYQQKP GKAPKLLIYK ASSLESGVPS RFSGSGSETH FTLTISSLQP DDVATYYCQ EYTSYGRTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPST LSASVGDRVT ITCRASQFIS RWLAWYQQKP GKAPKLLIYK ASSLESGVPS RFSGSGSETH FTLTISSLQP DDVATYYCQ EYTSYGRTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: Circumsporozoite protein
| Macromolecule | Name: Circumsporozoite protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.807488 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QEYQSYGSSS NTRVLNELNY DNAGTNLYNE LEMNYYGKQE NWYSLSSNSA SLGENDDGNN EDNEKLRKPK HKKLKQPADG NPDPNANPN VDPNANPNVD PNANPNVDPN ANPNANPNAN PNANPNANPN ANPNANPNAN PNANPNANPN ANPNANPNAN P NANPNANP ...String: QEYQSYGSSS NTRVLNELNY DNAGTNLYNE LEMNYYGKQE NWYSLSSNSA SLGENDDGNN EDNEKLRKPK HKKLKQPADG NPDPNANPN VDPNANPNVD PNANPNVDPN ANPNANPNAN PNANPNANPN ANPNANPNAN PNANPNANPN ANPNANPNAN P NANPNANP NANPNANPNV DPNANPNANP NANPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP NANPNANPNA NP NANPNAN PNANPNKNNQ GNGQGHNMPN DPNRNVDENA NANSAVKNNN NEEPSDKHIK EYLNKIQNSL STEWSPCSVT CGN GIQVRI KPGSANKPKD ELDYANDIEK KICKMEKCS UniProtKB: Circumsporozoite protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 63.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 45000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 147.9 |
|---|---|
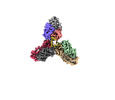
| Output model |  PDB-8ek1: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)