+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vigna radiata supercomplex I+III2 (full bridge) | |||||||||
 Map data Map data | Vigna radiata supercomplex I III2 (SC I III2) composite map of full supercomplex (bridged class 1). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | respiratory supercomplex / nadh-cyt c oxidoreductase / membrane complex / ELECTRON TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationTIM22 mitochondrial import inner membrane insertion complex / mitochondrial processing peptidase / P450-containing electron transport chain / photorespiration / protein insertion into mitochondrial inner membrane / NADH dehydrogenase complex / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / NADH dehydrogenase activity ...TIM22 mitochondrial import inner membrane insertion complex / mitochondrial processing peptidase / P450-containing electron transport chain / photorespiration / protein insertion into mitochondrial inner membrane / NADH dehydrogenase complex / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / NADH dehydrogenase activity / mitochondrial electron transport, ubiquinol to cytochrome c / electron transport coupled proton transport / acyl binding / NADH:ubiquinone reductase (H+-translocating) / acyl carrier activity / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / NADH dehydrogenase (ubiquinone) activity / quinone binding / ATP synthesis coupled electron transport / RNA splicing / aerobic respiration / chloroplast / respiratory electron transport chain / spliceosomal complex / electron transport chain / metalloendopeptidase activity / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / mRNA processing / 4 iron, 4 sulfur cluster binding / response to oxidative stress / electron transfer activity / mitochondrial inner membrane / nuclear speck / mitochondrial matrix / mRNA binding / heme binding / protein-containing complex binding / mitochondrion / proteolysis / metal ion binding / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Vigna radiata (mung bean) Vigna radiata (mung bean) | |||||||||
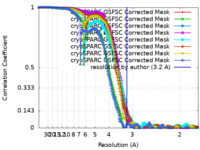
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Maldonado M / Letts JA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2023 Journal: Nat Plants / Year: 2023Title: Plant-specific features of respiratory supercomplex I + III from Vigna radiata. Authors: M Maldonado / Z Fan / K M Abe / J A Letts /  Abstract: The last steps of cellular respiration-an essential metabolic process in plants-are carried out by mitochondrial oxidative phosphorylation. This process involves a chain of multi-subunit membrane ...The last steps of cellular respiration-an essential metabolic process in plants-are carried out by mitochondrial oxidative phosphorylation. This process involves a chain of multi-subunit membrane protein complexes (complexes I-V) that form higher-order assemblies called supercomplexes. Although supercomplexes are the most physiologically relevant form of the oxidative phosphorylation complexes, their functions and structures remain mostly unknown. Here we present the cryogenic electron microscopy structure of the supercomplex I + III from Vigna radiata (mung bean). The structure contains the full subunit complement of complex I, including a newly assigned, plant-specific subunit. It also shows differences in the mitochondrial processing peptidase domain of complex III relative to a previously determined supercomplex with complex IV. The supercomplex interface, while reminiscent of that in other organisms, is plant specific, with a major interface involving complex III's mitochondrial processing peptidase domain and no participation of complex I's bridge domain. The complex I structure suggests that the bridge domain sets the angle between the enzyme's two arms, limiting large-scale conformational changes. Moreover, complex I's catalytic loops and its response in active-to-deactive assays suggest that, in V. radiata, the resting complex adopts a non-canonical state and can sample deactive- or open-like conformations even in the presence of substrate. This study widens our understanding of the possible conformations and behaviour of complex I and supercomplex I + III. Further studies of complex I and its supercomplexes in diverse organisms are needed to determine the universal and clade-specific mechanisms of respiration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27934.map.gz emd_27934.map.gz | 81.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27934-v30.xml emd-27934-v30.xml emd-27934.xml emd-27934.xml | 83.2 KB 83.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27934_fsc.xml emd_27934_fsc.xml emd_27934_fsc_2.xml emd_27934_fsc_2.xml emd_27934_fsc_3.xml emd_27934_fsc_3.xml emd_27934_fsc_4.xml emd_27934_fsc_4.xml emd_27934_fsc_5.xml emd_27934_fsc_5.xml emd_27934_fsc_6.xml emd_27934_fsc_6.xml emd_27934_fsc_7.xml emd_27934_fsc_7.xml emd_27934_fsc_8.xml emd_27934_fsc_8.xml | 19.9 KB 19.9 KB 19.9 KB 19.9 KB 19.9 KB 19.9 KB 20 KB 19.9 KB | Display Display Display Display Display Display Display Display |  FSC data file FSC data file |
| Images |  emd_27934.png emd_27934.png | 86.8 KB | ||
| Masks |  emd_27934_msk_1.map emd_27934_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27934.cif.gz emd-27934.cif.gz | 16.9 KB | ||
| Others |  emd_27934_additional_1.map.gz emd_27934_additional_1.map.gz emd_27934_half_map_1.map.gz emd_27934_half_map_1.map.gz emd_27934_half_map_2.map.gz emd_27934_half_map_2.map.gz | 779.1 MB 764 MB 764 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27934 http://ftp.pdbj.org/pub/emdb/structures/EMD-27934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27934 | HTTPS FTP |
-Related structure data
| Related structure data |  8e73MC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27934.map.gz / Format: CCP4 / Size: 87.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27934.map.gz / Format: CCP4 / Size: 87.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vigna radiata supercomplex I III2 (SC I III2) composite map of full supercomplex (bridged class 1). | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27934_msk_1.map emd_27934_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Vigna radiata supercomplex I III2 bridged class 1
| File | emd_27934_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vigna radiata supercomplex I III2 bridged class 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Vigna radiata supercomplex I III2 bridged class 1 half map B
| File | emd_27934_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vigna radiata supercomplex I III2 bridged class 1_half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Vigna radiata supercomplex I III2 bridged class 1 half map A
| File | emd_27934_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vigna radiata supercomplex I III2 bridged class 1_half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Vigna radiata supercomplex I+III2 (full bridge)
+Supramolecule #1: Vigna radiata supercomplex I+III2 (full bridge)
+Macromolecule #1: MPP-beta
+Macromolecule #2: MPP-alpha (protomer 1)
+Macromolecule #3: COB (cyt b)
+Macromolecule #4: CYC1 (cyt c1)
+Macromolecule #5: UCR1 (Rieske iron-sulfur protein subunit)
+Macromolecule #6: QCR7
+Macromolecule #7: QCR8
+Macromolecule #8: QCR6
+Macromolecule #9: QCR9
+Macromolecule #10: QCR10 (UCRY)
+Macromolecule #11: MPP-alpha (protomer 2)
+Macromolecule #12: Nad1
+Macromolecule #13: Nad2
+Macromolecule #14: Nad3
+Macromolecule #15: Nad4
+Macromolecule #16: Nad4L
+Macromolecule #17: Nad5
+Macromolecule #18: Nad6
+Macromolecule #19: NDUA1
+Macromolecule #20: NDUA2
+Macromolecule #21: NDUA3
+Macromolecule #22: NDUA5
+Macromolecule #23: NDUA6
+Macromolecule #24: NDUA7
+Macromolecule #25: NDUA8
+Macromolecule #26: NDUA9
+Macromolecule #27: NDUA11
+Macromolecule #28: NDUA12
+Macromolecule #29: NDUA13
+Macromolecule #30: NDUAB1-beta
+Macromolecule #31: NDUAB1-alpha
+Macromolecule #32: NDUB2
+Macromolecule #33: NDUB3
+Macromolecule #34: NDUB4
+Macromolecule #35: NDUB7
+Macromolecule #36: NDUB8
+Macromolecule #37: NDUB9
+Macromolecule #38: NDUB10
+Macromolecule #39: NDUB11
+Macromolecule #40: NDUFX
+Macromolecule #41: NDUC2
+Macromolecule #42: NDUP2
+Macromolecule #43: NDUCA1
+Macromolecule #44: NDUCA2
+Macromolecule #45: NDUCAL2
+Macromolecule #46: NDUS1
+Macromolecule #47: NDUS2
+Macromolecule #48: NDUS3
+Macromolecule #49: NDUS4
+Macromolecule #50: NDUS5
+Macromolecule #51: NDUS6
+Macromolecule #52: NDUS7
+Macromolecule #53: NDUS8
+Macromolecule #54: NDUP1
+Macromolecule #55: NDUP4
+Macromolecule #56: NDUB6
+Macromolecule #57: NDUV1
+Macromolecule #58: NDUV2
+Macromolecule #59: NDUX1
+Macromolecule #60: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #61: CARDIOLIPIN
+Macromolecule #62: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #63: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #64: HEME C
+Macromolecule #65: UBIQUINONE-10
+Macromolecule #66: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+Macromolecule #67: S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-b...
+Macromolecule #68: FE (III) ION
+Macromolecule #69: ZINC ION
+Macromolecule #70: IRON/SULFUR CLUSTER
+Macromolecule #71: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #72: FLAVIN MONONUCLEOTIDE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.55 mg/mL |
|---|---|
| Buffer | pH: 7.7 Details: 0.2% digitonin, 30 mM HEPES pH 7.7, 150 mM potassium acetate, 1 mM EDTA, 0.002% PMSF |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: LEICA EM GP / Details: 4 ul, 20 seconds pre-blot, blot 4 seconds. |
| Details | Digitonin-extracted, amphipol (A8-35)stabilized |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 25712 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8e73: |
 Movie
Movie Controller
Controller






































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)