[English] 日本語
 Yorodumi
Yorodumi- EMDB-27920: 3H03 Fab in complex with influenza virus neuraminidase from A/Bre... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3H03 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | influenza / monoclonal antibody / NA / H1N1 / cross-reactive antibody / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationexo-alpha-sialidase / exo-alpha-sialidase activity / viral budding from plasma membrane / carbohydrate metabolic process / host cell plasma membrane / virion membrane / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) / Influenza A virus (A/Brevig Mission/1/1918(H1N1)) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
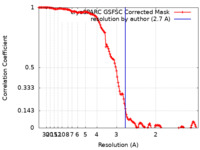
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Turner HL / Ozorowski G / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2023 Journal: Immunity / Year: 2023Title: Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo. Authors: Lena Hansen / Meagan McMahon / Hannah L Turner / Xueyong Zhu / Jackson S Turner / Gabriel Ozorowski / Daniel Stadlbauer / Juha Vahokoski / Aaron J Schmitz / Amena A Rizk / Wafaa B Alsoussi / ...Authors: Lena Hansen / Meagan McMahon / Hannah L Turner / Xueyong Zhu / Jackson S Turner / Gabriel Ozorowski / Daniel Stadlbauer / Juha Vahokoski / Aaron J Schmitz / Amena A Rizk / Wafaa B Alsoussi / Shirin Strohmeier / Wenli Yu / José Alberto Choreño-Parra / Luis Jiménez-Alvarez / Alfredo Cruz-Lagunas / Joaquín Zúñiga / Philip A Mudd / Rebecca J Cox / Ian A Wilson / Andrew B Ward / Ali H Ellebedy / Florian Krammer /    Abstract: Neuraminidase (NA) is one of the two influenza virus surface glycoproteins, and antibodies that target it are an independent correlate of protection. However, our current understanding of NA ...Neuraminidase (NA) is one of the two influenza virus surface glycoproteins, and antibodies that target it are an independent correlate of protection. However, our current understanding of NA antigenicity is incomplete. Here, we describe human monoclonal antibodies (mAbs) from a patient with a pandemic H1N1 virus infection in 2009. Two mAbs exhibited broad reactivity and inhibited NA enzyme activity of seasonal H1N1 viruses circulating before and after 2009, as well as viruses with avian or swine N1s. The mAbs provided robust protection from lethal challenge with human H1N1 and avian H5N1 viruses in mice, and both target an epitope on the lateral face of NA. In summary, we identified two broadly protective NA antibodies that share a novel epitope, inhibited NA activity, and provide protection against virus challenge in mice. Our work reaffirms that NA should be included as a target in future broadly protective or universal influenza virus vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27920.map.gz emd_27920.map.gz | 230.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27920-v30.xml emd-27920-v30.xml emd-27920.xml emd-27920.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27920_fsc.xml emd_27920_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_27920.png emd_27920.png | 116.5 KB | ||
| Masks |  emd_27920_msk_1.map emd_27920_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27920.cif.gz emd-27920.cif.gz | 6.7 KB | ||
| Others |  emd_27920_half_map_1.map.gz emd_27920_half_map_1.map.gz emd_27920_half_map_2.map.gz emd_27920_half_map_2.map.gz | 226 MB 226 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27920 http://ftp.pdbj.org/pub/emdb/structures/EMD-27920 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27920 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27920 | HTTPS FTP |
-Related structure data
| Related structure data |  8e6jMC  8e6kC  8eqaC  8eqcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27920.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27920.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.725 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27920_msk_1.map emd_27920_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_27920_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_27920_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 3H03 Fab in complex with influenza virus neuraminidase from A/Bre...
| Entire | Name: 3H03 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) |
|---|---|
| Components |
|
-Supramolecule #1: 3H03 Fab in complex with influenza virus neuraminidase from A/Bre...
| Supramolecule | Name: 3H03 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) Influenza A virus (A/Brevig Mission/1/1918(H1N1)) |
-Macromolecule #1: Neuraminidase
| Macromolecule | Name: Neuraminidase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: exo-alpha-sialidase |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) / Strain: A/Brevig Mission/1/1918 H1N1 Influenza A virus (A/Brevig Mission/1/1918(H1N1)) / Strain: A/Brevig Mission/1/1918 H1N1 |
| Molecular weight | Theoretical: 49.726387 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ADPHHHHHHS SSDYSDLQRV KQELLEEVKK ELQKVKEEII EAFVQELRKR GSLVPRGSPS RSVILTGNSS LCPISGWAIY SKDNGIRIG SKGDVFVIRE PFISCSHLEC RTFFLTQGAL LNDKHSNGTV KDRSPYRTLM SCPVGEAPSP YNSRFESVAW S ASACHDGM ...String: ADPHHHHHHS SSDYSDLQRV KQELLEEVKK ELQKVKEEII EAFVQELRKR GSLVPRGSPS RSVILTGNSS LCPISGWAIY SKDNGIRIG SKGDVFVIRE PFISCSHLEC RTFFLTQGAL LNDKHSNGTV KDRSPYRTLM SCPVGEAPSP YNSRFESVAW S ASACHDGM GWLTIGISGP DNGAVAVLKY NGIITDTIKS WRNNILRTQE SECACVNGSC FTIMTDGPSN GQASYKILKI EK GKVTKSI ELNAPNYHYE ECSCYPDTGK VMCVCRDNWH GSNRPWVSFD QNLDYQIGYI CSGVFGDNPR PNDGTGSCGP VSS NGANGI KGFSFRYDNG VWIGRTKSTS SRSGFEMIWD PNGWTETDSS FSVRQDIVAI TDWSGYSGSF VQHPELTGLD CMRP CFWVE LIRGQPKENT IWTSGSSISF CGVNSDTVGW SWPDGAELPF SIDK UniProtKB: Neuraminidase |
-Macromolecule #2: 3H03 fragment antigen binding light chain
| Macromolecule | Name: 3H03 fragment antigen binding light chain / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.361855 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSTSVGDRVT ITCRASQTIS TYLNWYQQKP GKAPELLIYV ASSLQSGVPS RFSGTGSGTE FTLTISSLQP GDFATYYCQ QSYSSPFTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPSS LSTSVGDRVT ITCRASQTIS TYLNWYQQKP GKAPELLIYV ASSLQSGVPS RFSGTGSGTE FTLTISSLQP GDFATYYCQ QSYSSPFTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: 3H03 fragment antigen binding heavy chain
| Macromolecule | Name: 3H03 fragment antigen binding heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.484332 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCTVSGDSIS SSYYYWGWIR QSPVKGLEWI GSFFYSGNTN YNPSLKSRVT ISVDTSKNQF SLNLRSVTA ADTAVYYCAR HVTSISSWNR GVYLDSWGRG ALVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN ...String: QVQLQESGPG LVKPSETLSL TCTVSGDSIS SSYYYWGWIR QSPVKGLEWI GSFFYSGNTN YNPSLKSRVT ISVDTSKNQF SLNLRSVTA ADTAVYYCAR HVTSISSWNR GVYLDSWGRG ALVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKRVEPKS C |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 3862 / Average exposure time: 7.51 sec. / Average electron dose: 41.1 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 190000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)