[English] 日本語
 Yorodumi
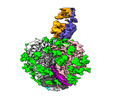
Yorodumi- EMDB-27735: Cryo-EM structure of SIVmac239 SOS-2P Env trimer in complex with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SIVmac239 SOS-2P Env trimer in complex with human bNAb PGT145 | |||||||||
 Map data Map data | sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CD4 / HIV-1 / SOSIP / Vaccine / IMMUNE SYSTEM / SIV / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion involved in viral entry into host cell / host cell endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||
| Biological species |  Simian immunodeficiency virus / Simian immunodeficiency virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.12 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Cryo-EM structures of prefusion SIV envelope trimer. Authors: Jason Gorman / Chunyan Wang / Rosemarie D Mason / Alexandra F Nazzari / Hugh C Welles / Tongqing Zhou / Julian W Bess / Tatsiana Bylund / Myungjin Lee / Yaroslav Tsybovsky / Raffaello ...Authors: Jason Gorman / Chunyan Wang / Rosemarie D Mason / Alexandra F Nazzari / Hugh C Welles / Tongqing Zhou / Julian W Bess / Tatsiana Bylund / Myungjin Lee / Yaroslav Tsybovsky / Raffaello Verardi / Shuishu Wang / Yongping Yang / Baoshan Zhang / Reda Rawi / Brandon F Keele / Jeffrey D Lifson / Jun Liu / Mario Roederer / Peter D Kwong /  Abstract: Simian immunodeficiency viruses (SIVs) are lentiviruses that naturally infect non-human primates of African origin and seeded cross-species transmissions of HIV-1 and HIV-2. Here we report prefusion ...Simian immunodeficiency viruses (SIVs) are lentiviruses that naturally infect non-human primates of African origin and seeded cross-species transmissions of HIV-1 and HIV-2. Here we report prefusion stabilization and cryo-EM structures of soluble envelope (Env) trimers from rhesus macaque SIV (SIV) in complex with neutralizing antibodies. These structures provide residue-level definition for SIV-specific disulfide-bonded variable loops (V1 and V2), which we used to delineate variable-loop coverage of the Env trimer. The defined variable loops enabled us to investigate assembled Env-glycan shields throughout SIV, which we found to comprise both N- and O-linked glycans, the latter emanating from V1 inserts, which bound the O-link-specific lectin jacalin. We also investigated in situ SIV-Env trimers on virions, determining cryo-electron tomography structures at subnanometer resolutions for an antibody-bound complex and a ligand-free state. Collectively, these structures define the prefusion-closed structure of the SIV-Env trimer and delineate variable-loop and glycan-shielding mechanisms of immune evasion conserved throughout SIV evolution. | |||||||||
| History |
|
- Structure visualization
Structure visualization
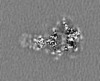
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27735.map.gz emd_27735.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27735-v30.xml emd-27735-v30.xml emd-27735.xml emd-27735.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27735.png emd_27735.png | 91.5 KB | ||
| Masks |  emd_27735_msk_1.map emd_27735_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27735.cif.gz emd-27735.cif.gz | 7.4 KB | ||
| Others |  emd_27735_additional_1.map.gz emd_27735_additional_1.map.gz emd_27735_additional_2.map.gz emd_27735_additional_2.map.gz emd_27735_half_map_1.map.gz emd_27735_half_map_1.map.gz emd_27735_half_map_2.map.gz emd_27735_half_map_2.map.gz | 44.5 MB 62 MB 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27735 http://ftp.pdbj.org/pub/emdb/structures/EMD-27735 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27735 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27735 | HTTPS FTP |
-Related structure data
| Related structure data |  8dvdMC  8duaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27735.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27735.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened | ||||||||||||||||||||||||||||||||||||
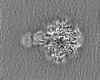
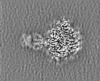
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27735_msk_1.map emd_27735_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: density modified (resolve)
| File | emd_27735_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modified (resolve) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened
| File | emd_27735_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_27735_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_27735_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SIVmac239 SOS-2P in complex with PGT145
| Entire | Name: SIVmac239 SOS-2P in complex with PGT145 |
|---|---|
| Components |
|
-Supramolecule #1: SIVmac239 SOS-2P in complex with PGT145
| Supramolecule | Name: SIVmac239 SOS-2P in complex with PGT145 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 16.822201 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GVFVLGFLGF LATAGSAMGA ASLTLTAQSR TLLAGIVQQQ QQLLDVPKRQ QELLRLPVWG TKNLQTRVTA IEKYLKDQAQ LNAWGCAFR QVCCTTVPWP NASLTPKWNN ETWQEWERKV DFLEENITAL LEEAQIQQEK NMYELQKLN UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 57.1955 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TLYVTVFYGV PAWRNATIPL FCATKNRDTW GTTQCLPDNG DYSEVALNVT ESFDAWNNTV TEQAIEDVWQ LFETSIKPCV KLSPLCITM RCNKSETDRW GLTKSITTTA STTSTTASAK VDMVNETSSC IAQDNCTGLE QEQMISCKFN MTGLKRDKTK E YNETWYSA ...String: TLYVTVFYGV PAWRNATIPL FCATKNRDTW GTTQCLPDNG DYSEVALNVT ESFDAWNNTV TEQAIEDVWQ LFETSIKPCV KLSPLCITM RCNKSETDRW GLTKSITTTA STTSTTASAK VDMVNETSSC IAQDNCTGLE QEQMISCKFN MTGLKRDKTK E YNETWYSA DLVCEQGNNT GNESRCYMNH CNTSVIQESC DKHYWDAIRF RYCAPPGYAL LRCNDTNYSG FMPKCSKVVV SS CTRMMET QTSTWFGFNG TRAENRTYIY WHGRDNRTII SLNKYYNLTM KCRRPGNKTV LPVTIMSGLV FHSQPINDRP KQA WCWFGG KWKDAIKEVK QTIVKHPRYT GTNNTDKINL TAPGGGDPEV TFMWTNCRGE FLYCKMNWFL NWVEDRNTAN QKPK EQHKR NYVPCHIRQI INTWHKVGKN VYLPPREGDL TCNSTVTSLI ANIDWIDGNQ TNITMSAEVA ELYRLELGDY KLVEI TPIG LAPTDCKRYT TGGTSR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: PGT145 Heavy
| Macromolecule | Name: PGT145 Heavy / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.393357 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGSSVKV SCKASGNSFS NHDVHWVRQA TGQGLEWMGW MSHEGDKTGL AQKFQGRVTI TRDSGASTVY MELRGLTAD DTAIYYCLTG SKHRLRDYFL (TYS)NE(TYS)GPNYEE WGDYLATLDV WGHGTAVTVS SASTKGPSVF PLA PSSKST ...String: QVQLVQSGAE VKKPGSSVKV SCKASGNSFS NHDVHWVRQA TGQGLEWMGW MSHEGDKTGL AQKFQGRVTI TRDSGASTVY MELRGLTAD DTAIYYCLTG SKHRLRDYFL (TYS)NE(TYS)GPNYEE WGDYLATLDV WGHGTAVTVS SASTKGPSVF PLA PSSKST SGGTAALGCL VKDYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNT KVDKK VEPKSCD |
-Macromolecule #4: PGT145 Light
| Macromolecule | Name: PGT145 Light / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.95375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVVITQSPLF LPVTPGEAAS LSCKCSHSLQ HSTGANYLAW YLQRPGQTPR LLIHLATHRA SGVPDRFSGS GSGTDFTLKI SRVESDDVG TYYCMQGLHS PWTFGQGTKV EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: EVVITQSPLF LPVTPGEAAS LSCKCSHSLQ HSTGANYLAW YLQRPGQTPR LLIHLATHRA SGVPDRFSGS GSGTDFTLKI SRVESDDVG TYYCMQGLHS PWTFGQGTKV EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Macromolecule #15: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 15 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #16: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 16 / Number of copies: 1 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 2.0 sec. / Average electron dose: 51.15 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)