+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CYP102A1 in Closed Conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cytochrome P450 electron transfer protein dynamic CYP102A1 / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationNADPH-hemoprotein reductase / NADPH-hemoprotein reductase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen / unspecific monooxygenase / aromatase activity / FMN binding / flavin adenine dinucleotide binding / iron ion binding / heme binding / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Su M / Xu H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Insight into the conformational dynamics of cytochrome P450 CYP102A1 enzyme using Cryo-EM Authors: Su M / Xu H | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27536.map.gz emd_27536.map.gz | 21 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27536-v30.xml emd-27536-v30.xml emd-27536.xml emd-27536.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27536.png emd_27536.png | 115.7 KB | ||
| Filedesc metadata |  emd-27536.cif.gz emd-27536.cif.gz | 6.4 KB | ||
| Others |  emd_27536_half_map_1.map.gz emd_27536_half_map_1.map.gz emd_27536_half_map_2.map.gz emd_27536_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27536 http://ftp.pdbj.org/pub/emdb/structures/EMD-27536 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27536 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27536 | HTTPS FTP |
-Validation report
| Summary document |  emd_27536_validation.pdf.gz emd_27536_validation.pdf.gz | 731.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27536_full_validation.pdf.gz emd_27536_full_validation.pdf.gz | 731.1 KB | Display | |
| Data in XML |  emd_27536_validation.xml.gz emd_27536_validation.xml.gz | 10 KB | Display | |
| Data in CIF |  emd_27536_validation.cif.gz emd_27536_validation.cif.gz | 11.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27536 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27536 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27536 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27536 | HTTPS FTP |
-Related structure data
| Related structure data |  8dmgMC  8dmeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27536.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27536.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.02 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27536_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27536_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CYP102A1
| Entire | Name: CYP102A1 |
|---|---|
| Components |
|
-Supramolecule #1: CYP102A1
| Supramolecule | Name: CYP102A1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: BM3 CYP102A |
|---|---|
| Source (natural) | Organism:  Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) |
| Molecular weight | Theoretical: 240 KDa |
-Macromolecule #1: Bifunctional cytochrome P450/NADPH--P450 reductase
| Macromolecule | Name: Bifunctional cytochrome P450/NADPH--P450 reductase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria)Strain: ATCC 14581 / DSM 32 / CCUG 1817 / JCM 2506 / NBRC 15308 / NCIMB 9376 / NCTC 10342 / NRRL B-14308 / VKM B-512 / Ford 19 |
| Molecular weight | Theoretical: 116.887188 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTIKEMPQPK TFGELKNLPL LNTDKPVQAL MKIADELGEI FKFEAPGRVT RYLSSQRLIK EACDESRFDK NLSQALKFVR DFFGDGLFT SWTHEKNWKK AHNILLPSFS QQAMKGYHAM MVDIAVQLVQ KWERLNADEH IEVPEDMTRL TLDTIGLCGF N YRFNSFYR ...String: MTIKEMPQPK TFGELKNLPL LNTDKPVQAL MKIADELGEI FKFEAPGRVT RYLSSQRLIK EACDESRFDK NLSQALKFVR DFFGDGLFT SWTHEKNWKK AHNILLPSFS QQAMKGYHAM MVDIAVQLVQ KWERLNADEH IEVPEDMTRL TLDTIGLCGF N YRFNSFYR DQPHPFITSM VRALDEAMNK LQRANPDDPA YDENKRQFQE DIKVMNDLVD KIIADRKASG EQSDDLLTHM LN GKDPETG EPLDDENIRY QIITFLIAGH ETTSGLLSFA LYFLVKNPHV LQKAAEEAAR VLVDPVPSYK QVKQLKYVGM VLN EALRLW PTAPAFSLYA KEDTVLGGEY PLEKGDELMV LIPQLHRDKT IWGDDVEEFR PERFENPSAI PQHAFKPFGN GQRA CIGQQ FALHEATLVL GMMLKHFDFE DHTNYELDIK ETLTLKPEGF VVKAKSKKIP LKKVRKKAEN AHNTPLLVLY GSNMG TAEG TARDLADIAM SKGFAPQVAT LDSHAGNLPR EGAVLIVTAS YNGHPPDNAK QFVDWLDQAS ADEVKGVRYS VFGCGD KNW ATTYQKVPAF IDETLAAKGA ENIADRGEAD ASDDFEGTYE EWREHMWSDV AAYFNLDIEN SEDNKSTLSL QFVDSAA DM PLAKMHGAFS TNVVASKELQ QPGSARSTRH LEIELPKEAS YQEGDHLGVI PRNYEGIVNR VTARFGLDAS QQIRLEAE E EKLAHLPLAK TVSVEELLQY VELQDPVTRT QLRAMAAKTV CPPHKVELEA LLEKQAYKEQ VLAKRLTMLE LLEKYPACE MKFSEFIALL PSIRPRYYSI SSSPRVDEKQ ASITVSVVSG EAWSGYGEYK GIASNYLAEL QEGDTITCFI STPQSEFTLP KDPETPLIM VGPGTGVAPF RGFVQARKQL KEQGQSLGEA HLYFGCRSPH EDYLYQEELE NAQSEGIITL HTAFSRMPNQ P KTYVQHVM EQDGKKLIEL LDQGAHFYIC GDGSQMAPAV EATLMKSYAD VHQVSEADAR LWLQQLEEKG RYAKDVWAG UniProtKB: Bifunctional cytochrome P450/NADPH--P450 reductase |
-Macromolecule #2: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 2 / Number of copies: 2 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #3: 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl...
| Macromolecule | Name: 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole type: ligand / ID: 3 / Number of copies: 2 / Formula: 1C6 |
|---|---|
| Molecular weight | Theoretical: 329.417 Da |
| Chemical component information |  ChemComp-1C6: |
-Macromolecule #4: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 4 / Number of copies: 2 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Macromolecule #5: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 5 / Number of copies: 2 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #6: SULFATE ION
| Macromolecule | Name: SULFATE ION / type: ligand / ID: 6 / Number of copies: 6 / Formula: SO4 |
|---|---|
| Molecular weight | Theoretical: 96.063 Da |
| Chemical component information |  ChemComp-SO4: |
-Macromolecule #7: TETRAETHYLENE GLYCOL
| Macromolecule | Name: TETRAETHYLENE GLYCOL / type: ligand / ID: 7 / Number of copies: 4 / Formula: PG4 |
|---|---|
| Molecular weight | Theoretical: 194.226 Da |
| Chemical component information |  ChemComp-PG4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 50.0 mM / Component - Formula: K2HPO4-KH2PO4-KCL / Component - Name: sodium phosphate + potassium chloride Details: Fresh prepare sodium phosphate (50mM) + potassium chloride (150mM), pH7.4 and filter with 0.22um filters. |
| Grid | Model: UltrAuFoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
| Details | The CYP102A1 sample purify and concentrated to 5mg/ml. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 15.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 249374 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



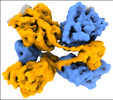




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)