[English] 日本語
 Yorodumi
Yorodumi- EMDB-27382: Helical rods of far-red light-absorbing allophycocyanin in Synech... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical rods of far-red light-absorbing allophycocyanin in Synechococcus sp. | ||||||||||||||||||
 Map data Map data | Sharpened and masked far-red light allophycocyanin | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology | Phycobilisome, alpha/beta subunit / Phycobilisome, alpha/beta subunit superfamily / Phycobilisome protein / phycobilisome / plasma membrane-derived thylakoid membrane / photosynthesis / Globin-like superfamily / Allophycocyanin subunit beta / Allophycocyanin subunit alpha Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  Synechococcus sp. 63AY4M1 (bacteria) Synechococcus sp. 63AY4M1 (bacteria) | ||||||||||||||||||
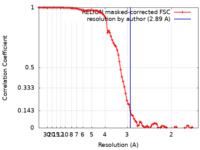
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | ||||||||||||||||||
 Authors Authors | Gisriel CJ / Shen GS / Soulier NT / Flesher DA / Brudvig GW / Bryant DA | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Helical allophycocyanin nanotubes absorb far-red light in a thermophilic cyanobacterium. Authors: Christopher J Gisriel / Eduard Elias / Gaozhong Shen / Nathan T Soulier / David A Flesher / M R Gunner / Gary W Brudvig / Roberta Croce / Donald A Bryant /   Abstract: To compete in certain low-light environments, some cyanobacteria express a paralog of the light-harvesting phycobiliprotein, allophycocyanin (AP), that strongly absorbs far-red light (FRL). Using ...To compete in certain low-light environments, some cyanobacteria express a paralog of the light-harvesting phycobiliprotein, allophycocyanin (AP), that strongly absorbs far-red light (FRL). Using cryo-electron microscopy and time-resolved absorption spectroscopy, we reveal the structure-function relationship of this FRL-absorbing AP complex (FRL-AP) that is expressed during acclimation to low light and that likely associates with chlorophyll a-containing photosystem I. FRL-AP assembles as helical nanotubes rather than typical toroids due to alterations of the domain geometry within each subunit. Spectroscopic characterization suggests that FRL-AP nanotubes are somewhat inefficient antenna; however, the enhanced ability to harvest FRL when visible light is severely attenuated represents a beneficial trade-off. The results expand the known diversity of light-harvesting proteins in nature and exemplify how biological plasticity is achieved by balancing resource accessibility with efficiency. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27382.map.gz emd_27382.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27382-v30.xml emd-27382-v30.xml emd-27382.xml emd-27382.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27382_fsc.xml emd_27382_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_27382.png emd_27382.png | 23.3 KB | ||
| Others |  emd_27382_additional_1.map.gz emd_27382_additional_1.map.gz emd_27382_half_map_1.map.gz emd_27382_half_map_1.map.gz emd_27382_half_map_2.map.gz emd_27382_half_map_2.map.gz | 54.2 MB 54.5 MB 54.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27382 http://ftp.pdbj.org/pub/emdb/structures/EMD-27382 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27382 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27382 | HTTPS FTP |
-Related structure data
| Related structure data |  8ddyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27382.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27382.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened and masked far-red light allophycocyanin | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened
| File | emd_27382_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half1
| File | emd_27382_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half2
| File | emd_27382_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Far-red light allophycocyanin
| Entire | Name: Far-red light allophycocyanin |
|---|---|
| Components |
|
-Supramolecule #1: Far-red light allophycocyanin
| Supramolecule | Name: Far-red light allophycocyanin / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Synechococcus sp. 63AY4M1 (bacteria) Synechococcus sp. 63AY4M1 (bacteria) |
-Macromolecule #1: Allophycocyanin subunit alpha
| Macromolecule | Name: Allophycocyanin subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Synechococcus sp. 63AY4M1 (bacteria) Synechococcus sp. 63AY4M1 (bacteria) |
| Molecular weight | Theoretical: 20.526518 KDa |
| Recombinant expression | Organism:  Synechococcus sp. PCC 7002 (bacteria) Synechococcus sp. PCC 7002 (bacteria) |
| Sequence | String: MGHHHHHHHH HHSSGHIEGR HMQAAASMSI VAQVIAQSDA ADRFLSSAEI AKLEDFFSKG QVRIRAAQKL AENEQKIVQE GSKRFWAKC PNTPSNKGNP QKTALCQRDQ GWYIRLVSYC ILAGNDKPLE DIGLNGMREM YISLGVPLPN LRVAMSCLKE V AAGILSSE EMALAAPYFD RLIRAF |
-Macromolecule #2: Allophycocyanin subunit beta
| Macromolecule | Name: Allophycocyanin subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Synechococcus sp. 63AY4M1 (bacteria) Synechococcus sp. 63AY4M1 (bacteria) |
| Molecular weight | Theoretical: 17.712303 KDa |
| Recombinant expression | Organism:  Synechococcus sp. PCC 7002 (bacteria) Synechococcus sp. PCC 7002 (bacteria) |
| Sequence | String: MKDTITSLIN PADEKGSYLD AAALEQLNRY FQSGNMRVKA AKTISSSASS IISKTVAKSL LYGDITLPGG (MEN)MYPTR RYA ACLRDLTYFL RYATYAMLAA DPSILDERVL QGLKETYITL GVPIDRVIQA LNAMKEVLTE SLDTEASQEM AVYLDHI IA GLS |
-Macromolecule #3: PHYCOCYANOBILIN
| Macromolecule | Name: PHYCOCYANOBILIN / type: ligand / ID: 3 / Number of copies: 6 / Formula: CYC |
|---|---|
| Molecular weight | Theoretical: 588.694 Da |
| Chemical component information |  ChemComp-CYC: |
-Macromolecule #4: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 4 / Number of copies: 3 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 330 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 59.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)