+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structures of bAE1 captured in multiple states. | |||||||||
 Map data Map data | Main final full map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryoEM / Band3 / bAE1 (SLC4A1) / anion exchanger / STRUCTURAL PROTEIN / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmonoatomic anion transmembrane transporter activity / solute:inorganic anion antiporter activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Zhekova HR / Wang WG / Jiang JS / Tsirulnikov K / Muhammad-Khan GH / Azimov R / Abuladze N / Kao L / Newman D / Noskov SY ...Zhekova HR / Wang WG / Jiang JS / Tsirulnikov K / Muhammad-Khan GH / Azimov R / Abuladze N / Kao L / Newman D / Noskov SY / Teleman P / Zhou ZH / Pushkin A / Kurtz I | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations. Authors: Hristina R Zhekova / Jiansen Jiang / Weiguang Wang / Kirill Tsirulnikov / Gülru Kayık / Hanif Muhammad Khan / Rustam Azimov / Natalia Abuladze / Liyo Kao / Debbie Newman / Sergei Yu Noskov ...Authors: Hristina R Zhekova / Jiansen Jiang / Weiguang Wang / Kirill Tsirulnikov / Gülru Kayık / Hanif Muhammad Khan / Rustam Azimov / Natalia Abuladze / Liyo Kao / Debbie Newman / Sergei Yu Noskov / D Peter Tieleman / Z Hong Zhou / Alexander Pushkin / Ira Kurtz /   Abstract: Anion exchanger 1 (AE1, band 3) is a major membrane protein of red blood cells and plays a key role in acid-base homeostasis, urine acidification, red blood cell shape regulation, and removal of ...Anion exchanger 1 (AE1, band 3) is a major membrane protein of red blood cells and plays a key role in acid-base homeostasis, urine acidification, red blood cell shape regulation, and removal of carbon dioxide during respiration. Though structures of the transmembrane domain (TMD) of three SLC4 transporters, including AE1, have been resolved previously in their outward-facing (OF) state, no mammalian SLC4 structure has been reported in the inward-facing (IF) conformation. Here we present the cryoEM structures of full-length bovine AE1 with its TMD captured in both IF and OF conformations. Remarkably, both IF-IF homodimers and IF-OF heterodimers were detected. The IF structures feature downward movement in the core domain with significant unexpected elongation of TM11. Molecular modeling and structure guided mutagenesis confirmed the functional significance of residues involved in TM11 elongation. Our data provide direct evidence for an elevator-like mechanism of ion transport by an SLC4 family member. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27267.map.gz emd_27267.map.gz | 24.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27267-v30.xml emd-27267-v30.xml emd-27267.xml emd-27267.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27267.png emd_27267.png | 80.4 KB | ||
| Filedesc metadata |  emd-27267.cif.gz emd-27267.cif.gz | 6.5 KB | ||
| Others |  emd_27267_additional_1.map.gz emd_27267_additional_1.map.gz emd_27267_additional_2.map.gz emd_27267_additional_2.map.gz emd_27267_half_map_1.map.gz emd_27267_half_map_1.map.gz emd_27267_half_map_2.map.gz emd_27267_half_map_2.map.gz | 23.9 MB 5.8 MB 19.5 MB 19.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27267 http://ftp.pdbj.org/pub/emdb/structures/EMD-27267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27267 | HTTPS FTP |
-Validation report
| Summary document |  emd_27267_validation.pdf.gz emd_27267_validation.pdf.gz | 720.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27267_full_validation.pdf.gz emd_27267_full_validation.pdf.gz | 719.8 KB | Display | |
| Data in XML |  emd_27267_validation.xml.gz emd_27267_validation.xml.gz | 10.4 KB | Display | |
| Data in CIF |  emd_27267_validation.cif.gz emd_27267_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27267 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27267 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27267 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27267 | HTTPS FTP |
-Related structure data
| Related structure data |  8d9nMC  8e34C  8eeqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27267.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27267.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main final full map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
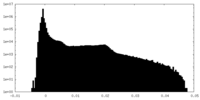
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Partially connected CD.
| File | emd_27267_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Partially connected CD. | ||||||||||||
| Projections & Slices |
| ||||||||||||
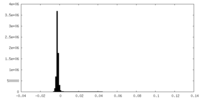
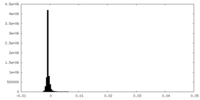
| Density Histograms |
-Additional map: Fully connected CD.
| File | emd_27267_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Fully connected CD. | ||||||||||||
| Projections & Slices |
| ||||||||||||
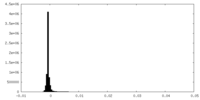
| Density Histograms |
-Half map: half1 map.
| File | emd_27267_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
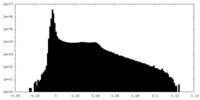
| Density Histograms |
-Half map: half2 map.
| File | emd_27267_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
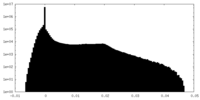
| Density Histograms |
- Sample components
Sample components
-Entire : band 3 anion transport protein
| Entire | Name: band 3 anion transport protein |
|---|---|
| Components |
|
-Supramolecule #1: band 3 anion transport protein
| Supramolecule | Name: band 3 anion transport protein / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104 KDa |
-Macromolecule #1: Anion exchange protein
| Macromolecule | Name: Anion exchange protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104.474258 KDa |
| Sequence | String: MGDPEEYEDQ LEETLEQKEY EDHDSVSIPM EEAEGDTIQE EEAEARVNQL TDTDYHTTSQ HPETHKVCVQ LRELVMDEKN QEIQWMETA RWVGLEENLG KDGIWGRPHL PYLNFWSLLE LQKAFAKGTV LLDLPGKSLA EVANQLLDRF TFEGQIQPDD Q DNLLRVLL ...String: MGDPEEYEDQ LEETLEQKEY EDHDSVSIPM EEAEGDTIQE EEAEARVNQL TDTDYHTTSQ HPETHKVCVQ LRELVMDEKN QEIQWMETA RWVGLEENLG KDGIWGRPHL PYLNFWSLLE LQKAFAKGTV LLDLPGKSLA EVANQLLDRF TFEGQIQPDD Q DNLLRVLL LKHSHASDME ALGGVKPVVV THSGDPSEPL LPQHPSLETE LFCEQGEGST RGHAPEILGK SPQDWEATLV LV GCARFLK RPVLGFVRLK EPMEPEPKPE GSEEPAVPVR FLIVLLGPEG PNINYTQLGR AAATLMSERV FWNDAYLAQS KET LVQSLE GFLDCSLVLP PLDAPSEKAL LSLVPVQKEL LRRRYLPSPA KPDPSIFKDL DVKKGPGDTP EDPLQRTGKL FGGL VRDIR RRYPRYLSDI TDALSPQVLS AIIFIYFAAL TPAITFGGLL GDKTENMIGV SELLLSTALQ GIIFSLLGAQ PLLVL GFSG PLLVFEEAFY SFCQTNNLEY IVGRVWIGFW LILLVVLVVA FEGSFLVRFI SRYTQEIFSF LISLIFIYET FYKLVT IFQ DHPLQKNYDH DVLTTPKPQA ALPNTALLSL VLMAGTFFLA MMLRKFKNSS YFPGKLRRII GDFGVPISIL IMVMVDA LI QDTYTQKLSV PEGLSVSNPT ERDWLIHPLG IRVEFPIWMM FASALPALLV FILIFLESQI TTLIISKPER KMVKGSGF H LDLLLIIGMG GVGAIFGMPW LSATTVRTVT HANALTVMSK DSTPGAVSQI QGVKEQRISG LLVAVLVGVS ILMGPVLRH IPLAVLFGIF LYMGVTSLSG IQLFDRVLLL LKPRKYYPEV PYARRVKTWR MHLFTITQIV CLVVLWVVRS IKQISLALPF ILILTVPLR RFLLPFIFRD MELKLLDADD VKLNLDEQNG QDEYDEVAMP V UniProtKB: Anion exchange protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-48 / Average exposure time: 12.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 36764 / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.4000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8d9n: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)