[English] 日本語
 Yorodumi
Yorodumi- EMDB-27020: Human glycogenin-1 and glycogen synthase-1 complex in the presenc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human glycogenin-1 and glycogen synthase-1 complex in the presence of glucose-6-phosphate | |||||||||
 Map data Map data | Sharpened map of GYS1:GYG1 complex in the presence of G6P | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Metabolism Glycogen synthesis / Glycogen synthase Glycogenin / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglycogen synthase activity, transferring glucose-1-phosphate / Glycogen storage disease type XV (GYG1) / Glycogen storage disease type 0 (muscle GYS1) / glycogen(starch) synthase / glycogenin glucosyltransferase / Glycogen storage disease type II (GAA) / glycogenin glucosyltransferase activity / alpha-1,4-glucan glucosyltransferase (UDP-glucose donor) activity / D-glucose binding / glycogen biosynthetic process ...glycogen synthase activity, transferring glucose-1-phosphate / Glycogen storage disease type XV (GYG1) / Glycogen storage disease type 0 (muscle GYS1) / glycogen(starch) synthase / glycogenin glucosyltransferase / Glycogen storage disease type II (GAA) / glycogenin glucosyltransferase activity / alpha-1,4-glucan glucosyltransferase (UDP-glucose donor) activity / D-glucose binding / glycogen biosynthetic process / Glycogen breakdown (glycogenolysis) / glycosyltransferase activity / inclusion body / Myoclonic epilepsy of Lafora / Glycogen synthesis / lysosomal lumen / manganese ion binding / heart development / secretory granule lumen / ficolin-1-rich granule lumen / Neutrophil degranulation / protein homodimerization activity / extracellular region / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
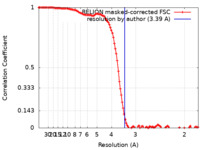
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Liu Y / Fastman NM / Tzitzilonis C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: The structural mechanism of human glycogen synthesis by the GYS1-GYG1 complex. Authors: Nathan M Fastman / Yuxi Liu / Vyas Ramanan / Hanne Merritt / Eileen Ambing / Anna A DePaoli-Roach / Peter J Roach / Thomas D Hurley / Kevin T Mellem / Julie C Ullman / Eric Green / David ...Authors: Nathan M Fastman / Yuxi Liu / Vyas Ramanan / Hanne Merritt / Eileen Ambing / Anna A DePaoli-Roach / Peter J Roach / Thomas D Hurley / Kevin T Mellem / Julie C Ullman / Eric Green / David Morgans / Christos Tzitzilonis /  Abstract: Glycogen is the primary energy reserve in mammals, and dysregulation of glycogen metabolism can result in glycogen storage diseases (GSDs). In muscle, glycogen synthesis is initiated by the enzymes ...Glycogen is the primary energy reserve in mammals, and dysregulation of glycogen metabolism can result in glycogen storage diseases (GSDs). In muscle, glycogen synthesis is initiated by the enzymes glycogenin-1 (GYG1), which seeds the molecule by autoglucosylation, and glycogen synthase-1 (GYS1), which extends the glycogen chain. Although both enzymes are required for proper glycogen production, the nature of their interaction has been enigmatic. Here, we present the human GYS1:GYG1 complex in multiple conformations representing different functional states. We observe an asymmetric conformation of GYS1 that exposes an interface for close GYG1 association, and propose this state facilitates handoff of the GYG1-associated glycogen chain to a GYS1 subunit for elongation. Full activation of GYS1 widens the GYG1-binding groove, enabling GYG1 release concomitant with glycogen chain growth. This structural mechanism connecting chain nucleation and extension explains the apparent stepwise nature of glycogen synthesis and suggests distinct states to target for GSD-modifying therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27020.map.gz emd_27020.map.gz | 122.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27020-v30.xml emd-27020-v30.xml emd-27020.xml emd-27020.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27020_fsc.xml emd_27020_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27020.png emd_27020.png | 167.7 KB | ||
| Masks |  emd_27020_msk_1.map emd_27020_msk_1.map | 129.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27020.cif.gz emd-27020.cif.gz | 6.7 KB | ||
| Others |  emd_27020_additional_1.map.gz emd_27020_additional_1.map.gz emd_27020_half_map_1.map.gz emd_27020_half_map_1.map.gz emd_27020_half_map_2.map.gz emd_27020_half_map_2.map.gz | 64.4 MB 120.6 MB 120.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27020 http://ftp.pdbj.org/pub/emdb/structures/EMD-27020 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27020 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27020 | HTTPS FTP |
-Related structure data
| Related structure data |  8cvxMC  8cvyC  8cvzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27020.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27020.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of GYS1:GYG1 complex in the presence of G6P | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9142 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27020_msk_1.map emd_27020_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
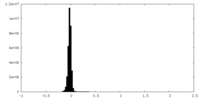
| Density Histograms |
-Additional map: Unsharpened of GYS1:GYG1 complex in the presence of G6P
| File | emd_27020_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened of GYS1:GYG1 complex in the presence of G6P | ||||||||||||
| Projections & Slices |
| ||||||||||||
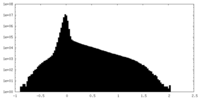
| Density Histograms |
-Half map: Half map A of GYS1:GYG1 complex in the presence of G6P
| File | emd_27020_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of GYS1:GYG1 complex in the presence of G6P | ||||||||||||
| Projections & Slices |
| ||||||||||||
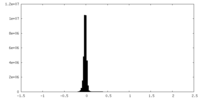
| Density Histograms |
-Half map: Half map B of GYS1:GYG1 complex in the presence of G6P
| File | emd_27020_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of GYS1:GYG1 complex in the presence of G6P | ||||||||||||
| Projections & Slices |
| ||||||||||||
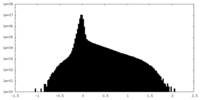
| Density Histograms |
- Sample components
Sample components
-Entire : Human glycogenin-1 and glycogen synthase-1 complex in the presenc...
| Entire | Name: Human glycogenin-1 and glycogen synthase-1 complex in the presence of glucose-6-phosphate |
|---|---|
| Components |
|
-Supramolecule #1: Human glycogenin-1 and glycogen synthase-1 complex in the presenc...
| Supramolecule | Name: Human glycogenin-1 and glycogen synthase-1 complex in the presence of glucose-6-phosphate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: Glycogen [starch] synthase, muscle
| Macromolecule | Name: Glycogen [starch] synthase, muscle / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: glycogen(starch) synthase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.634438 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPLNRTLEMS ELPGLEDWED EFDLENAVLF EVAWEVANKV GGIYTVLQTK AKVTGDEWGD NYFLVGPYTE QGVRTQVELL EAPTPALKR TLDSMNSKGC KVYFGRWLIE GGPLVVLLDV GASAWALERW KGELWDTCNI GVPWYDREAN DAVLFGFLTT W FLGEFLAQ ...String: MPLNRTLEMS ELPGLEDWED EFDLENAVLF EVAWEVANKV GGIYTVLQTK AKVTGDEWGD NYFLVGPYTE QGVRTQVELL EAPTPALKR TLDSMNSKGC KVYFGRWLIE GGPLVVLLDV GASAWALERW KGELWDTCNI GVPWYDREAN DAVLFGFLTT W FLGEFLAQ SEEKPHVVAH FHEWLAGVGL CLCRARRLPV ATIFTTHATL LGRYLCAGAV DFYNNLENFN VDKEAGERQI YH RYCMERA AAHCAHVFTT VSQITAIEAQ HLLKRKPDIV TPNGLNVKKF SAMHEFQNLH AQSKARIQEF VRGHFYGHLD FNL DKTLYF FIAGRYEFSN KGADVFLEAL ARLNYLLRVN GSEQTVVAFF IMPARTNNFN VETLKGQAVR KQLWDTANTV KEKF GRKLY ESLLVGSLPD MNKMLDKEDF TMMKRAIFAT QRQSFPPVCT HNMLDDSSDP ILTTIRRIGL FNSSADRVKV IFHPE FLSS TSPLLPVDYE EFVRGCHLGV FPSYYEPWGY TPAECTVMGI PSISTNLSGF GCFMEEHIAD PSAYGIYILD RRFRSL DDS CSQLTSFLYS FCQQSRRQRI IQRNRTERLS DLLDWKYLGR YYMSARHMAL SKAFPEHFTY EPNEADAAQG Y UniProtKB: Glycogen [starch] synthase, muscle |
-Macromolecule #2: Glycogenin-1
| Macromolecule | Name: Glycogenin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: glycogenin glucosyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.560789 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPMTDQAFVT LTTNDAYAKG ALVLGSSLKQ HRTTRRLVVL ATPQVSDSMR KVLETVFDEV IMVDVLDSGD SAHLTLMKRP ELGVTLTKL HCWSLTQYSK CVFMDADTLV LANIDDLFDR EELSAAPDPG WPDCFNSGVF VYQPSVETYN QLLHLASEQG S FDGGDQGI ...String: GPMTDQAFVT LTTNDAYAKG ALVLGSSLKQ HRTTRRLVVL ATPQVSDSMR KVLETVFDEV IMVDVLDSGD SAHLTLMKRP ELGVTLTKL HCWSLTQYSK CVFMDADTLV LANIDDLFDR EELSAAPDPG WPDCFNSGVF VYQPSVETYN QLLHLASEQG S FDGGDQGI LNTFFSSWAT TDIRKHLPFI YNLSSISIFS YLPAFKVFGA SAKVVHFLGR VKPWNYTYDP KTKSVKSEAH DP NMTHPEF LILWWNIFTT NVLPLLQQFG LVKDTCSYVN VLSDLVYTLA FSCGFCRKED VSGAISHLSL GEIPAMAQPF VSS EERKER WEQGQADYMG ADSFDNIKRK LDTYLQ UniProtKB: Glycogenin-1 |
-Macromolecule #3: 6-O-phosphono-alpha-D-glucopyranose
| Macromolecule | Name: 6-O-phosphono-alpha-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: G6P |
|---|---|
| Molecular weight | Theoretical: 260.136 Da |
| Chemical component information | 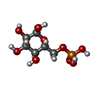 ChemComp-G6P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 9 sec blotting time, 15 blot force. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 6797 / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8cvx: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)