+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
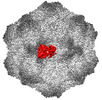
| Title | CPV Affinity Purified Polyclonal Fab A Site Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CPV / polyclonal Fab / A site / vaccination / VIRUS / VIRUS-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont entry into host cell via permeabilization of host membrane / microtubule-dependent intracellular transport of viral material towards nucleus / adhesion receptor-mediated virion attachment to host cell / T=1 icosahedral viral capsid / viral penetration into host nucleus / host cell / clathrin-dependent endocytosis of virus by host cell / entry receptor-mediated virion attachment to host cell / host cell nucleus / structural molecule activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Hartmann SR / Hafenstein SL / Charnesky AJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Cryo EM structures map a post vaccination polyclonal antibody response to canine parvovirus. Authors: Samantha R Hartmann / Andrew J Charnesky / Simon P Früh / Robert A López-Astacio / Wendy S Weichert / Nadia DiNunno / Sung Hung Cho / Carol M Bator / Colin R Parrish / Susan L Hafenstein /  Abstract: Canine parvovirus (CPV) is an important pathogen that emerged by cross-species transmission to cause severe disease in dogs. To understand the host immune response to vaccination, sera from dogs ...Canine parvovirus (CPV) is an important pathogen that emerged by cross-species transmission to cause severe disease in dogs. To understand the host immune response to vaccination, sera from dogs immunized with parvovirus are obtained, the polyclonal antibodies are purified and used to solve the high resolution cryo EM structures of the polyclonal Fab-virus complexes. We use a custom software, Icosahedral Subparticle Extraction and Correlated Classification (ISECC) to perform subparticle analysis and reconstruct polyclonal Fab-virus complexes from two different dogs eight and twelve weeks post vaccination. In the resulting polyclonal Fab-virus complexes there are a total of five distinct Fabs identified. In both cases, any of the five antibodies identified would interfere with receptor binding. This polyclonal mapping approach identifies a specific, limited immune response to the live vaccine virus and allows us to investigate the binding of multiple different antibodies or ligands to virus capsids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26786.map.gz emd_26786.map.gz | 26.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26786-v30.xml emd-26786-v30.xml emd-26786.xml emd-26786.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26786_fsc.xml emd_26786_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_26786.png emd_26786.png | 206.7 KB | ||
| Filedesc metadata |  emd-26786.cif.gz emd-26786.cif.gz | 5.9 KB | ||
| Others |  emd_26786_additional_1.map.gz emd_26786_additional_1.map.gz emd_26786_additional_2.map.gz emd_26786_additional_2.map.gz emd_26786_half_map_1.map.gz emd_26786_half_map_1.map.gz emd_26786_half_map_2.map.gz emd_26786_half_map_2.map.gz | 777.3 MB 405.4 MB 23.1 MB 23.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26786 http://ftp.pdbj.org/pub/emdb/structures/EMD-26786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26786 | HTTPS FTP |
-Validation report
| Summary document |  emd_26786_validation.pdf.gz emd_26786_validation.pdf.gz | 830.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26786_full_validation.pdf.gz emd_26786_full_validation.pdf.gz | 830.3 KB | Display | |
| Data in XML |  emd_26786_validation.xml.gz emd_26786_validation.xml.gz | 12.6 KB | Display | |
| Data in CIF |  emd_26786_validation.cif.gz emd_26786_validation.cif.gz | 17.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26786 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26786 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26786 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26786 | HTTPS FTP |
-Related structure data
| Related structure data |  7utpMC  7utrC  7utsC  7utuC  7utvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26786.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26786.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #2
| File | emd_26786_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
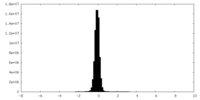
| Density Histograms |
-Additional map: #1
| File | emd_26786_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
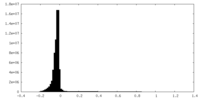
| Density Histograms |
-Half map: #2
| File | emd_26786_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
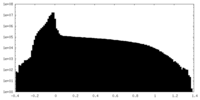
| Density Histograms |
-Half map: #1
| File | emd_26786_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
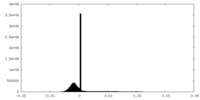
| Density Histograms |
- Sample components
Sample components
-Entire : Canine parvovirus in complex with polyclonal fab
| Entire | Name: Canine parvovirus in complex with polyclonal fab |
|---|---|
| Components |
|
-Supramolecule #1: Canine parvovirus in complex with polyclonal fab
| Supramolecule | Name: Canine parvovirus in complex with polyclonal fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.562367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GVGISTGTFN NQTEFKFLEN GWVEITANSS RLVHLNMPES ENYRRVVVNN MDKTAVNGNM ALDDIHAQIV TPWSLVDANA WGVWFNPGD WQLIVNTMSE LHLVSFEQEI FNVVLKTVSE SATQPPTKVY NNDLTASLMV ALDSNNTMPF TPAAMRSETL G FYPWKPTI ...String: GVGISTGTFN NQTEFKFLEN GWVEITANSS RLVHLNMPES ENYRRVVVNN MDKTAVNGNM ALDDIHAQIV TPWSLVDANA WGVWFNPGD WQLIVNTMSE LHLVSFEQEI FNVVLKTVSE SATQPPTKVY NNDLTASLMV ALDSNNTMPF TPAAMRSETL G FYPWKPTI PTPWRYYFQW DRTLIPSHTG TSGTPTNIYH GTDPDDVQFY TIENSVPVHL LRTGDEFATG TFFFDCKPCR LT HTWQTNR ALGLPPFLNS LPQSEGATNF GDIGVQQDKR RGVTQMGNTN YITEATIMRP AEVGYSAPYY SFEASTQGPF KTP IAAGRG GAQTDENQAA DGNPRYAFGR QHGQKTTTTG ETPERFTYIA HQDTGRYPEG DWIQNINFNL PVTNDNVLLP TDPI GGKTG INYTNIFNTY GPLTALNNVP PVYPNGQIWD KEFDTDLKPR LHVNAPFVCQ NNCPGQLFVK VAPNLTNEYD PDASA NMSR IVTYSDFWWK GKLVFKAKLR ASHTWNPIQQ MSINVDNQFN YVPSNIGGMK IVYEKSQLAP RKLY UniProtKB: Capsid protein VP1 |
-Macromolecule #2: Light chain antibody fragment
| Macromolecule | Name: Light chain antibody fragment / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.443399 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK) |
-Macromolecule #3: Heavy chain antibody fragment
| Macromolecule | Name: Heavy chain antibody fragment / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.209344 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)