+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2676 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of bovine ComplexI | |||||||||
 Map data Map data | Reconstruction of bovine ComplexI | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NADH dehydrogenase / respiratory complex | |||||||||
| Biological species |  | |||||||||
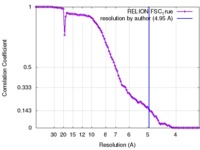
| Method | single particle reconstruction / cryo EM / Resolution: 4.95 Å | |||||||||
 Authors Authors | Vinothkumar KR / Zhu J / Hirst J | |||||||||
 Citation Citation |  Journal: Nature / Year: 2014 Journal: Nature / Year: 2014Title: Architecture of mammalian respiratory complex I. Authors: Kutti R Vinothkumar / Jiapeng Zhu / Judy Hirst /  Abstract: Complex I (NADH:ubiquinone oxidoreductase) is essential for oxidative phosphorylation in mammalian mitochondria. It couples electron transfer from NADH to ubiquinone with proton translocation across ...Complex I (NADH:ubiquinone oxidoreductase) is essential for oxidative phosphorylation in mammalian mitochondria. It couples electron transfer from NADH to ubiquinone with proton translocation across the energy-transducing inner membrane, providing electrons for respiration and driving ATP synthesis. Mammalian complex I contains 44 different nuclear- and mitochondrial-encoded subunits, with a combined mass of 1 MDa. The 14 conserved 'core' subunits have been structurally defined in the minimal, bacterial complex, but the structures and arrangement of the 30 'supernumerary' subunits are unknown. Here we describe a 5 Å resolution structure of complex I from Bos taurus heart mitochondria, a close relative of the human enzyme, determined by single-particle electron cryo-microscopy. We present the structures of the mammalian core subunits that contain eight iron-sulphur clusters and 60 transmembrane helices, identify 18 supernumerary transmembrane helices, and assign and model 14 supernumerary subunits. Thus, we considerably advance knowledge of the structure of mammalian complex I and the architecture of its supernumerary ensemble around the core domains. Our structure provides insights into the roles of the supernumerary subunits in regulation, assembly and homeostasis, and a basis for understanding the effects of mutations that cause a diverse range of human diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2676.map.gz emd_2676.map.gz | 78.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2676-v30.xml emd-2676-v30.xml emd-2676.xml emd-2676.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2676_fsc.xml emd_2676_fsc.xml | 8.1 KB | Display |  FSC data file FSC data file |
| Images |  ComplexI_image_forEMDB.tif ComplexI_image_forEMDB.tif | 377.5 KB | ||
| Others |  emd_2676_half_map_1.map.gz emd_2676_half_map_1.map.gz emd_2676_half_map_2.map.gz emd_2676_half_map_2.map.gz | 65.5 MB 65.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2676 http://ftp.pdbj.org/pub/emdb/structures/EMD-2676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2676 | HTTPS FTP |
-Related structure data
| Related structure data |  4uq8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2676.map.gz / Format: CCP4 / Size: 81.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2676.map.gz / Format: CCP4 / Size: 81.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of bovine ComplexI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.717 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 2676 half map 1.map
| File | emd_2676_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 2676 half map 2.map
| File | emd_2676_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NADH:ubiquinone oxidoreductase
| Entire | Name: NADH:ubiquinone oxidoreductase |
|---|---|
| Components |
|
-Supramolecule #1000: NADH:ubiquinone oxidoreductase
| Supramolecule | Name: NADH:ubiquinone oxidoreductase / type: sample / ID: 1000 Details: The enzyme was isolated from bovine heart mitochondria in detergent micelles and imaged on ice as single particles Oligomeric state: 1 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1 MDa / Theoretical: 1 MDa |
-Macromolecule #1: NADH:ubiquinone oxidoreductase
| Macromolecule | Name: NADH:ubiquinone oxidoreductase / type: protein_or_peptide / ID: 1 / Name.synonym: Complex I / Number of copies: 1 / Oligomeric state: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 1 MDa / Theoretical: 1 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20mM Tris-HCl, 150 mM NaCl, 0.03% Cymal-7 |
| Grid | Details: 300 mesh holey-carbon Quantifoil gold grid R 0.6/1 glow discharged in air |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 100 K / Instrument: OTHER Method: The specimen was vitrified in an environmental plunge-freeze apparatus (Bellare et al, J.Electr. Micros. Tech., 1988, 10, 87-111). Blot for 15-18 seconds after the diameter of the blotted ...Method: The specimen was vitrified in an environmental plunge-freeze apparatus (Bellare et al, J.Electr. Micros. Tech., 1988, 10, 87-111). Blot for 15-18 seconds after the diameter of the blotted meniscus ceases to expand and plunged. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80 K / Max: 90 K / Average: 85 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 120,000 times magnification once at the start of data collection |
| Details | Exposure intensity set to give 50 electron/pixel/second at the detector, which translates to ~17 electrons/square_Angstrom/second at the specimen. Each image was exposed for 4 seconds. |
| Date | Oct 21, 2013 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Digitization - Sampling interval: 14 µm / Number real images: 1154 / Average electron dose: 64 e/Å2 Details: An in-house built system was used to intercept the frames from the detector at a rate of 18 frames per second. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 81495 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)