[English] 日本語
 Yorodumi
Yorodumi- EMDB-26650: Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruva... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruvate dehydrogenase complex from bovine kidney | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | e2 / pyruvate / dehydrogenase / complex / TRANSFERASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationSignaling by Retinoic Acid / PDH complex synthesizes acetyl-CoA from PYR / Regulation of pyruvate dehydrogenase (PDH) complex / : / Protein lipoylation / pyruvate catabolic process / dihydrolipoyllysine-residue acetyltransferase / dihydrolipoyllysine-residue acetyltransferase activity / pyruvate dehydrogenase complex / tricarboxylic acid cycle ...Signaling by Retinoic Acid / PDH complex synthesizes acetyl-CoA from PYR / Regulation of pyruvate dehydrogenase (PDH) complex / : / Protein lipoylation / pyruvate catabolic process / dihydrolipoyllysine-residue acetyltransferase / dihydrolipoyllysine-residue acetyltransferase activity / pyruvate dehydrogenase complex / tricarboxylic acid cycle / glucose metabolic process / mitochondrial matrix / identical protein binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
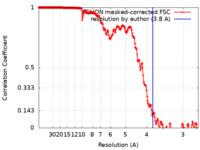
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||
 Authors Authors | Liu S / Xia X / Zhen J / Li ZH / Zhou ZH | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2022 Journal: Cell Discov / Year: 2022Title: Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney. Authors: Shiheng Liu / Xian Xia / James Zhen / Zihang Li / Z Hong Zhou /  Abstract: The α-keto acid dehydrogenase complex family catalyzes the essential oxidative decarboxylation of α-keto acids to yield acyl-CoA and NADH. Despite performing the same overarching reaction, members ...The α-keto acid dehydrogenase complex family catalyzes the essential oxidative decarboxylation of α-keto acids to yield acyl-CoA and NADH. Despite performing the same overarching reaction, members of the family have different component structures and structural organization between each other and across phylogenetic species. While native structures of α-keto acid dehydrogenase complexes from bacteria and fungi became available recently, the atomic structure and organization of their mammalian counterparts in native states remain unknown. Here, we report the cryo-electron microscopy structures of the endogenous cubic 2-oxoglutarate dehydrogenase complex (OGDC) and icosahedral pyruvate dehydrogenase complex (PDC) cores from bovine kidney determined at resolutions of 3.5 Å and 3.8 Å, respectively. The structures of multiple proteins were reconstructed from a single lysate sample, allowing direct structural comparison without the concerns of differences arising from sample preparation and structure determination. Although native and recombinant E2 core scaffold structures are similar, the native structures are decorated with their peripheral E1 and E3 subunits. Asymmetric sub-particle reconstructions support heterogeneity in the arrangements of these peripheral subunits. In addition, despite sharing a similar monomeric fold, OGDC and PDC E2 cores have distinct interdomain and intertrimer interactions, which suggests a means of modulating self-assembly to mitigate heterologous binding between mismatched E2 species. The lipoyl moiety lies near a mobile gatekeeper within the interdomain active site of OGDC E2 and PDC E2. Analysis of the twofold related intertrimer interface identified secondary structural differences and chemical interactions between icosahedral and cubic geometries of the core. Taken together, our study provides a direct structural comparison of OGDC and PDC from the same source and offers new insights into determinants of interdomain interactions and of architecture diversity among α-keto acid dehydrogenase complexes. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26650.map.gz emd_26650.map.gz | 666 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26650-v30.xml emd-26650-v30.xml emd-26650.xml emd-26650.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26650_fsc.xml emd_26650_fsc.xml | 20.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_26650.png emd_26650.png | 160.2 KB | ||
| Masks |  emd_26650_msk_1.map emd_26650_msk_1.map | 729 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26650.cif.gz emd-26650.cif.gz | 5.9 KB | ||
| Others |  emd_26650_half_map_1.map.gz emd_26650_half_map_1.map.gz emd_26650_half_map_2.map.gz emd_26650_half_map_2.map.gz | 581.7 MB 581.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26650 http://ftp.pdbj.org/pub/emdb/structures/EMD-26650 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26650 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26650 | HTTPS FTP |
-Validation report
| Summary document |  emd_26650_validation.pdf.gz emd_26650_validation.pdf.gz | 752.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26650_full_validation.pdf.gz emd_26650_full_validation.pdf.gz | 752.3 KB | Display | |
| Data in XML |  emd_26650_validation.xml.gz emd_26650_validation.xml.gz | 28.2 KB | Display | |
| Data in CIF |  emd_26650_validation.cif.gz emd_26650_validation.cif.gz | 38 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26650 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26650 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26650 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26650 | HTTPS FTP |
-Related structure data
| Related structure data |  7uomMC  7uolC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26650.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26650.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26650_msk_1.map emd_26650_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26650_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26650_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruva...
| Entire | Name: Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruvate dehydrogenase complex from bovine kidney |
|---|---|
| Components |
|
-Supramolecule #1: Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruva...
| Supramolecule | Name: Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruvate dehydrogenase complex from bovine kidney type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69 KDa |
-Macromolecule #1: Acetyltransferase component of pyruvate dehydrogenase complex
| Macromolecule | Name: Acetyltransferase component of pyruvate dehydrogenase complex type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO / EC number: dihydrolipoyllysine-residue acetyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69.142594 KDa |
| Sequence | String: MWRVCARRAQ NAAPRAGFGA RWTAFREEPG APCVTPQAGS ALARCSSKTP GYGRVRALCG WSPVSRATPR NRVLLQLWGS PSRRWYSLP PHQKVPLPSL SPTMQAGTIA RWEKKEGEKI NEGELIAEVE TDKATVGFES VEECYMAKIL VAEGTRDVPV G AIICITVD ...String: MWRVCARRAQ NAAPRAGFGA RWTAFREEPG APCVTPQAGS ALARCSSKTP GYGRVRALCG WSPVSRATPR NRVLLQLWGS PSRRWYSLP PHQKVPLPSL SPTMQAGTIA RWEKKEGEKI NEGELIAEVE TDKATVGFES VEECYMAKIL VAEGTRDVPV G AIICITVD KPEDVEAFKN YTLDSSAAPA PPAAPAPTPA APAPSPTPSA QAPGSSYPTH MQVLLPALSP TMTMGTVQRW EK KVGEKLN EGDLLAEIET DKATIGFEVQ EEGYLAKILI PEGTRDVPLG TPLCIIVEKE ADIPAFADYR PAEVTDLKPP APP PIPSPA APVPPAPQPV APPPSAPRPA APAGPKGRVF VSPLAKKLAA EKGIDLTQVK GTGPDGRIIK KDIDSFVPTK AAPT PAAAV PPPSPGVAPV PTGVFTDIPI SNIRRVIAQR LMQSKQTIPH YYLSIDVNMG EVLLVRKELN KMLEGKSKIS VNDFI IKAS ALACLKVPEA NSSWMDTVIR QNHVVDISVA VSTPAGLITP IVFNAHIKGL ETIANDVVSL ATKAREGKLQ PHEFQG GTF TISNLGMFGI KNFSAIINPP QACILAIGAS EDRLVPADNE KGFDVASMMS VTLSCDHRVV DGAVGAQWLA EFRKYLE KP ITMLL UniProtKB: Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 8.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Software | Name: UCSF ChimeraX |
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
| Output model |  PDB-7uom: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)