+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of LARGE1 from C1 reconstruction | ||||||||||||
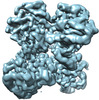
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Glycosyltransferase / Metalloenzyme / TRANSFERASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpost-embryonic hindlimb morphogenesis / Defective LARGE causes MDDGA6 and MDDGB6 / xylosyltransferase activity / principal sensory nucleus of trigeminal nerve development / walking behavior / connective tissue development / glycosphingolipid biosynthetic process / O-linked glycosylation / skeletal muscle organ development / Transferases; Glycosyltransferases ...post-embryonic hindlimb morphogenesis / Defective LARGE causes MDDGA6 and MDDGB6 / xylosyltransferase activity / principal sensory nucleus of trigeminal nerve development / walking behavior / connective tissue development / glycosphingolipid biosynthetic process / O-linked glycosylation / skeletal muscle organ development / Transferases; Glycosyltransferases / glucuronosyltransferase activity / UDP-xylosyltransferase activity / synaptic assembly at neuromuscular junction / localization of cell / protein O-linked glycosylation via mannose / reactive gliosis / N-acetylglucosamine metabolic process / glycoprotein biosynthetic process / neuromuscular process controlling posture / acetylglucosaminyltransferase activity / water transport / retina layer formation / plasma membrane organization / retina vasculature development in camera-type eye / skeletal muscle fiber differentiation / nerve development / basement membrane organization / hexosyltransferase activity / neuromuscular synaptic transmission / dentate gyrus development / skeletal muscle tissue regeneration / protein O-linked glycosylation / astrocyte differentiation / cardiac muscle cell development / acetylcholine receptor signaling pathway / protein targeting to membrane / Transferases; Glycosyltransferases; Hexosyltransferases / muscle cell cellular homeostasis / : / blood vessel development / glycosyltransferase activity / response to light stimulus / macrophage differentiation / Transferases; Glycosyltransferases; Pentosyltransferases / response to mechanical stimulus / behavioral fear response / striated muscle contraction / skeletal muscle fiber development / potassium ion transmembrane transport / myelination / cytoskeleton organization / post-translational protein modification / determination of adult lifespan / protein localization to plasma membrane / intracellular protein transport / neuromuscular junction / sensory perception of sound / bone development / multicellular organism growth / memory / neuron migration / long-term synaptic potentiation / manganese ion binding / protein-containing complex assembly / gene expression / Golgi membrane / Golgi apparatus / protein-containing complex / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Joseph S / Schnicker NJ / Campbell KP | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of LARGE1 from C1 reconstruction Authors: Campbell KP | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26540.map.gz emd_26540.map.gz | 91.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26540-v30.xml emd-26540-v30.xml emd-26540.xml emd-26540.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26540.png emd_26540.png | 196.5 KB | ||
| Filedesc metadata |  emd-26540.cif.gz emd-26540.cif.gz | 5.8 KB | ||
| Others |  emd_26540_half_map_1.map.gz emd_26540_half_map_1.map.gz emd_26540_half_map_2.map.gz emd_26540_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26540 http://ftp.pdbj.org/pub/emdb/structures/EMD-26540 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26540 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26540 | HTTPS FTP |
-Related structure data
| Related structure data |  7ui6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26540.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26540.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8015 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_26540_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26540_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : LARGE1
| Entire | Name: LARGE1 |
|---|---|
| Components |
|
-Supramolecule #1: LARGE1
| Supramolecule | Name: LARGE1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: No transmembrane region |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 211 KDa |
-Macromolecule #1: Xylosyl- and glucuronyltransferase LARGE1
| Macromolecule | Name: Xylosyl- and glucuronyltransferase LARGE1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: Transferases; Glycosyltransferases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.806242 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSALLILALV GAAVADYKDH DGDYKDHDID YKDDDDKLAA AFEDGKPVSL SPLESQAHSP RYTASSQRER ESLEVRMREV EEENRALRR QLSLAQGRAP SHRRGNHSKT YSMEEGTGDS ENLRAGIVAG NSSECGQQPV VEKCETIHVA IVCAGYNASR D VVTLVKSV ...String: MSALLILALV GAAVADYKDH DGDYKDHDID YKDDDDKLAA AFEDGKPVSL SPLESQAHSP RYTASSQRER ESLEVRMREV EEENRALRR QLSLAQGRAP SHRRGNHSKT YSMEEGTGDS ENLRAGIVAG NSSECGQQPV VEKCETIHVA IVCAGYNASR D VVTLVKSV LFHRRNPLHF HLIADSIAEQ ILATLFQTWM VPAVRVDFYN ADELKSEVSW IPNKHYSGIY GLMKLVLTKT LP ANLERVI VLDTDITFAT DIAELWAVFH KFKGQQVLGL VENQSDWYLG NLWKNHRPWP ALGRGYNTGV ILLLLDKLRK MKW EQMWRL TAERELMGML STSLADQDIF NAVIKQNPFL VYQLPCFWNV QLSDHTRSEQ CYRDVSDLKV IHWNSPKKLR VKNK HVEFF RNLYLTFLEY DGNLLRRELF GCPSEADVNS ENLQKQLSEL DEDDLCYEFR RERFTVHRTH LYFLHYEYEP AADST DVTL VAQLSMDRLQ MLEAICKHWE GPISLALYLS DAEAQQFLRY AQGSEVLMSR HNVGYHIVYK EGQFYPVNLL RNVAMK HIS TPYMFLSDID FLPMYGLYEY LRKSVIQLDL ANTKKAMIVP AFETLRYRLS FPKSKAELLS MLDMGTLFTF RYHVWTK GH APTNFAKWRT ATTPYRVEWE ADFEPYVVVR RDCPEYDRRF VGFGWNKVAH IMELDVQEYE FIVLPNAYMI HMPHAPSF D ITKFRSNKQY RICLKTLKEE FQQDMSRRYG FAALKYLTAE NNSHHHHHH UniProtKB: Xylosyl- and glucuronyltransferase LARGE1 |
-Macromolecule #2: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 6.6 |
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 1.664 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Used initial model from Alphafold2 |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-7ui6: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)