+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of porcine kidney V-ATPase with SidK, Rotary State 3 | |||||||||||||||
 Map data Map data | Map used for model building. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | proton translocation / complex / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationROS and RNS production in phagocytes / RHOA GTPase cycle / Transferrin endocytosis and recycling / Amino acids regulate mTORC1 / Ion channel transport / plasma membrane proton-transporting V-type ATPase complex / Insulin receptor recycling / symbiont-mediated suppression of host phagosome acidification / eye pigmentation / central nervous system maturation ...ROS and RNS production in phagocytes / RHOA GTPase cycle / Transferrin endocytosis and recycling / Amino acids regulate mTORC1 / Ion channel transport / plasma membrane proton-transporting V-type ATPase complex / Insulin receptor recycling / symbiont-mediated suppression of host phagosome acidification / eye pigmentation / central nervous system maturation / rostrocaudal neural tube patterning / proton-transporting V-type ATPase, V1 domain / positive regulation of transforming growth factor beta1 production / proton-transporting two-sector ATPase complex, catalytic domain / synaptic vesicle lumen acidification / vacuolar transport / proton-transporting V-type ATPase, V0 domain / cellular response to increased oxygen levels / vacuolar proton-transporting V-type ATPase, V1 domain / vacuolar proton-transporting V-type ATPase, V0 domain / endosome to plasma membrane protein transport / clathrin-coated vesicle membrane / lysosomal lumen acidification / proton-transporting V-type ATPase complex / head morphogenesis / vacuolar proton-transporting V-type ATPase complex / osteoclast development / vacuolar acidification / regulation of cellular pH / dendritic spine membrane / vacuolar membrane / microvillus / ATPase activator activity / regulation of MAPK cascade / autophagosome membrane / proton-transporting ATPase activity, rotational mechanism / positive regulation of Wnt signaling pathway / transporter activator activity / ATP metabolic process / H+-transporting two-sector ATPase / transport vesicle / angiotensin maturation / RNA endonuclease activity / proton transmembrane transport / transmembrane transport / small GTPase binding / synaptic vesicle membrane / melanosome / positive regulation of canonical Wnt signaling pathway / signaling receptor activity / presynapse / ATPase binding / intracellular iron ion homeostasis / early endosome / lysosome / endosome / endosome membrane / apical plasma membrane / external side of plasma membrane / lysosomal membrane / endoplasmic reticulum membrane / ATP hydrolysis activity / ATP binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||
 Authors Authors | Tan YZ | |||||||||||||||
| Funding support |  Canada, Canada,  Singapore, 4 items Singapore, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2022 Journal: Life Sci Alliance / Year: 2022Title: CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7. Authors: Yong Zi Tan / Yazan M Abbas / Jing Ze Wu / Di Wu / Kristine A Keon / Geoffrey G Hesketh / Stephanie A Bueler / Anne-Claude Gingras / Carol V Robinson / Sergio Grinstein / John L Rubinstein /   Abstract: V-ATPases are rotary proton pumps that serve as signaling hubs with numerous protein binding partners. CryoEM with exhaustive focused classification allowed detection of endogenous proteins ...V-ATPases are rotary proton pumps that serve as signaling hubs with numerous protein binding partners. CryoEM with exhaustive focused classification allowed detection of endogenous proteins associated with porcine kidney V-ATPase. An extra C subunit was found in ∼3% of complexes, whereas ∼1.6% of complexes bound mEAK-7, a protein with proposed roles in dauer formation in nematodes and mTOR signaling in mammals. High-resolution cryoEM of porcine kidney V-ATPase with recombinant mEAK-7 showed that mEAK-7's TLDc domain interacts with V-ATPase's stator, whereas its C-terminal α helix binds V-ATPase's rotor. This crosslink would be expected to inhibit rotary catalysis. However, unlike the yeast TLDc protein Oxr1p, exogenous mEAK-7 does not inhibit V-ATPase and mEAK-7 overexpression in cells does not alter lysosomal or phagosomal pH. Instead, cryoEM suggests that the mEAK-7:V-ATPase interaction is disrupted by ATP-induced rotation of the rotor. Comparison of Oxr1p and mEAK-7 binding explains this difference. These results show that V-ATPase binding by TLDc domain proteins can lead to effects ranging from strong inhibition to formation of labile interactions that are sensitive to the enzyme's activity. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26388.map.gz emd_26388.map.gz | 12 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26388-v30.xml emd-26388-v30.xml emd-26388.xml emd-26388.xml | 35.6 KB 35.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26388_fsc.xml emd_26388_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_26388.png emd_26388.png | 49.4 KB | ||
| Filedesc metadata |  emd-26388.cif.gz emd-26388.cif.gz | 9.9 KB | ||
| Others |  emd_26388_additional_1.map.gz emd_26388_additional_1.map.gz emd_26388_half_map_1.map.gz emd_26388_half_map_1.map.gz emd_26388_half_map_2.map.gz emd_26388_half_map_2.map.gz | 97.4 MB 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26388 http://ftp.pdbj.org/pub/emdb/structures/EMD-26388 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26388 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26388 | HTTPS FTP |
-Related structure data
| Related structure data |  7u8rMC  7u8oC  7u8pC  7u8qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-10874 (Title: Single-Particle CryoEM of mammalian V-ATPase with the TLDc domain protein mEAK7 bound (Various Datasets) EMPIAR-10874 (Title: Single-Particle CryoEM of mammalian V-ATPase with the TLDc domain protein mEAK7 bound (Various Datasets)Data size: 12.7 TB Data #1: Unaligned multiframe movies of Pig Kidney V-ATPase bound to mEAK-7 collected using Tundra [micrographs - multiframe] Data #2: Aligned and dose-weighted micrographs of Pig Kidney V-ATPase bound to mEAK-7 collected using Tundra [micrographs - single frame] Data #3: Polished particles of Pig Kidney V-ATPase bound to mEAK-7 collected using Tundra [picked particles - single frame - processed] Data #4: Unaligned multiframe movies of Pig Kidney V-ATPase bound to mEAK-7 collected using Titan Krios and Falcon4 [micrographs - multiframe] Data #5: Aligned and dose-weighted micrographs of Pig Kidney V-ATPase bound to mEAK-7 collected using Titan Krios and Falcon4 [micrographs - single frame] Data #6: Polished particles of Pig Kidney V-ATPase bound to mEAK-7 collected using Titan Krios and Falcon4 [picked particles - multiframe - processed] Data #7: Unaligned multiframe movies of Pig Kidney V-ATPase bound to mEAK-7deltaCterm collected using Titan Krios and Falcon4 [micrographs - multiframe] Data #8: Aligned and dose-weighted micrographs of Pig Kidney V-ATPase bound to mEAK-7deltaCterm collected using Titan Krios and Falcon4 [micrographs - single frame] Data #9: Polished particles of Pig Kidney V-ATPase bound to mEAK-7deltaCterm collected using Titan Krios and Falcon4 [picked particles - single frame - processed] Data #10: Unaligned multiframe movies of Pig Kidney V-ATPase bound to mEAK-7 with ATP collected using Titan Krios and Falcon4 [micrographs - multiframe] Data #11: Aligned and dose-weighted micrographs of Pig Kidney V-ATPase bound to mEAK-7 with ATP collected using Titan Krios and Falcon4 [micrographs - single frame] Data #12: Polished particles of Pig Kidney V-ATPase bound to mEAK-7 with ATP collected using Titan Krios and Falcon4 [picked particles - single frame - processed] Data #13: Unaligned multiframe movies of Pig Kidney V-ATPase bound to mEAK-7 with EDTA/EGTA collected using Titan Krios and Falcon4 [micrographs - multiframe] Data #14: Aligned and dose-weighted micrographs of Pig Kidney V-ATPase bound to mEAK-7 with EDTA/EGTA collected using Titan Krios and Falcon4 [micrographs - single frame] Data #15: Polished particles of Pig Kidney V-ATPase bound to mEAK-7 with EDTA/EGTA collected using Titan Krios and Falcon4 [picked particles - single frame - processed] Data #16: Unaligned multiframe movies of Pig Kidney V-ATPase bound to mEAK-7 with Calcium collected using Glacios with Selectris X and Falcon 4 [micrographs - multiframe] Data #17: Aligned and dose-weighted micrographs of Pig Kidney V-ATPase bound to mEAK-7 with Calcium collected using Glacios with Selectris X and Falcon 4 [micrographs - single frame] Data #18: Polished particles of Pig Kidney V-ATPase bound to mEAK-7 with Calcium collected using Glacios with Selectris X and Falcon 4 [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26388.map.gz / Format: CCP4 / Size: 13.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26388.map.gz / Format: CCP4 / Size: 13.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map used for model building. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.52801 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: V-ATPase with SidK, Rotary State 3
| File | emd_26388_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | V-ATPase with SidK, Rotary State 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
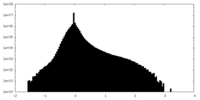
| Density Histograms |
-Half map: V-ATPase with SidK, Rotary State 3
| File | emd_26388_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | V-ATPase with SidK, Rotary State 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
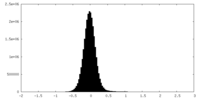
| Density Histograms |
-Half map: V-ATPase with SidK, Rotary State 3
| File | emd_26388_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | V-ATPase with SidK, Rotary State 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
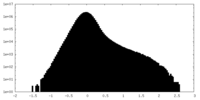
| Density Histograms |
- Sample components
Sample components
+Entire : Porcine kidney V-ATPase with SidK, Rotary State 3
+Supramolecule #1: Porcine kidney V-ATPase with SidK, Rotary State 3
+Macromolecule #1: V-type proton ATPase catalytic subunit A
+Macromolecule #2: Vacuolar proton pump subunit B
+Macromolecule #3: V-type proton ATPase subunit C
+Macromolecule #4: V-type proton ATPase subunit D
+Macromolecule #5: V-type proton ATPase subunit E 1
+Macromolecule #6: V-type proton ATPase subunit F
+Macromolecule #7: V-type proton ATPase subunit G
+Macromolecule #8: Bacterial effector protein SidK
+Macromolecule #9: V-type proton ATPase subunit H
+Macromolecule #10: V-type proton ATPase subunit a
+Macromolecule #11: V-type proton ATPase 21 kDa proteolipid subunit isoform 1
+Macromolecule #12: ATPase H+ transporting accessory protein 1
+Macromolecule #13: V-type proton ATPase subunit
+Macromolecule #14: V-type proton ATPase subunit
+Macromolecule #15: Ribonuclease kappa
+Macromolecule #16: V-type proton ATPase proteolipid subunit
+Macromolecule #17: ATPase H(+)-transporting lysosomal accessory protein 2
+Macromolecule #18: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.911445 µm / Nominal defocus min: 0.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)