+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Influenza Neuraminidase N1-MI15-sNAp-174 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza / Antigen / Engineered Protein / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN | |||||||||
| Biological species |   Influenza A virus Influenza A virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.25 Å | |||||||||
 Authors Authors | Acton OJ / Veesler D / Seattle Structural Genomics Center for Infectious Disease (SSGCID) | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure-based design of stabilized recombinant influenza neuraminidase tetramers. Authors: Daniel Ellis / Julia Lederhofer / Oliver J Acton / Yaroslav Tsybovsky / Sally Kephart / Christina Yap / Rebecca A Gillespie / Adrian Creanga / Audrey Olshefsky / Tyler Stephens / Deleah ...Authors: Daniel Ellis / Julia Lederhofer / Oliver J Acton / Yaroslav Tsybovsky / Sally Kephart / Christina Yap / Rebecca A Gillespie / Adrian Creanga / Audrey Olshefsky / Tyler Stephens / Deleah Pettie / Michael Murphy / Claire Sydeman / Maggie Ahlrichs / Sidney Chan / Andrew J Borst / Young-Jun Park / Kelly K Lee / Barney S Graham / David Veesler / Neil P King / Masaru Kanekiyo /  Abstract: Influenza virus neuraminidase (NA) is a major antiviral drug target and has recently reemerged as a key target of antibody-mediated protective immunity. Here we show that recombinant NAs across non- ...Influenza virus neuraminidase (NA) is a major antiviral drug target and has recently reemerged as a key target of antibody-mediated protective immunity. Here we show that recombinant NAs across non-bat subtypes adopt various tetrameric conformations, including an "open" state that may help explain poorly understood variations in NA stability across viral strains and subtypes. We use homology-directed protein design to uncover the structural principles underlying these distinct tetrameric conformations and stabilize multiple recombinant NAs in the "closed" state, yielding two near-atomic resolution structures of NA by cryo-EM. In addition to enhancing thermal stability, conformational stabilization improves affinity to protective antibodies elicited by viral infection, including antibodies targeting a quaternary epitope and the broadly conserved catalytic site. Stabilized NAs can also be integrated into viruses without affecting fitness. Our findings provide a deeper understanding of NA structure, stability, and antigenicity, and establish design strategies for reinforcing the conformational integrity of recombinant NA proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26319.map.gz emd_26319.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26319-v30.xml emd-26319-v30.xml emd-26319.xml emd-26319.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26319.png emd_26319.png | 68.5 KB | ||
| Filedesc metadata |  emd-26319.cif.gz emd-26319.cif.gz | 5.8 KB | ||
| Others |  emd_26319_additional_1.map.gz emd_26319_additional_1.map.gz | 16.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26319 http://ftp.pdbj.org/pub/emdb/structures/EMD-26319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26319 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26319.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26319.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
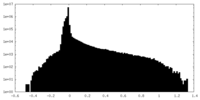
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_26319_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
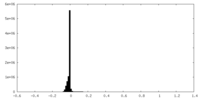
| Density Histograms |
- Sample components
Sample components
-Entire : Structure-based design of an engineered Influenza Neuraminidase t...
| Entire | Name: Structure-based design of an engineered Influenza Neuraminidase tetramer |
|---|---|
| Components |
|
-Supramolecule #1: Structure-based design of an engineered Influenza Neuraminidase t...
| Supramolecule | Name: Structure-based design of an engineered Influenza Neuraminidase tetramer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 210 kDa/nm |
-Macromolecule #1: Influenza N1-MI15-sNAp-174
| Macromolecule | Name: Influenza N1-MI15-sNAp-174 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 42.577742 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VKLAGNSSLC PVSGWAPLSK DNSVRIGSKG DVFVIREPFI SCSPLECRQF FLTQGALLND KHSNGTIKDR SPYRTLMSVP IGSVPSPYN ARFESIAWSA SACHDGINWL TIGITGPDSG AVAILKYNGI ITDTIKSWRN NILRTQESEC ACVNGSCFTI M TDGPSDGQ ...String: VKLAGNSSLC PVSGWAPLSK DNSVRIGSKG DVFVIREPFI SCSPLECRQF FLTQGALLND KHSNGTIKDR SPYRTLMSVP IGSVPSPYN ARFESIAWSA SACHDGINWL TIGITGPDSG AVAILKYNGI ITDTIKSWRN NILRTQESEC ACVNGSCFTI M TDGPSDGQ ASYKIFRIEK GKIIKSVEMK APNYHYEECS CYPDSSEITC VCRDNWHGSN RPWVSFNQNL EYQMGYICSG VF GDNPRPN DKTGSCGPVS SNGANGVKGF SFKYGNGVWI GRTKSISSRK GFEMIWDPNG WTGTDNKFSI KQDIVGINEW SGY SGSFVM HPELTGLDCI VPCFWVELIR GRPEENTIWT SGSSISFCGV NSDTVGWSWP DGAELPFTID K |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C4 (4 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.25 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 72106 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)