[English] 日本語
 Yorodumi
Yorodumi- EMDB-25909: Adeno-associated Virus Go.1 at 2.9 Angstroms resolution, AAVGo.1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Adeno-associated Virus Go.1 at 2.9 Angstroms resolution, AAVGo.1 AAV-Go | |||||||||
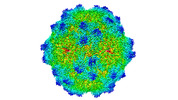
 Map data Map data | Cryo-EM map of AAV-Go.1 virus like particle at 2.9 Angstroms resolution. Pixel size of 0.661 Angstroms. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAVGo.1 / AAV / AAV-Go / AAVR / adeno-associated virus / PKD domain / parvovirus / virus / gene therapy / receptor / adeno-associated virus receptor / PKD1 / PKD / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP1/VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   Adeno-associated virus Adeno-associated virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.93 Å | |||||||||
 Authors Authors | Silveria M / Large E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2022 Journal: J Virol / Year: 2022Title: Cross-Species Permissivity: Structure of a Goat Adeno-Associated Virus and Its Complex with the Human Receptor AAVR. Authors: Edward E Large / Mark A Silveria / Onellah Weerakoon / Tommi A White / Michael S Chapman /  Abstract: Adeno-associated virus (AAV) is a small ssDNA satellite virus of high interest (in recombinant form) as a safe and effective gene therapy vector. AAV's human cell entry receptor (AAVR) contains ...Adeno-associated virus (AAV) is a small ssDNA satellite virus of high interest (in recombinant form) as a safe and effective gene therapy vector. AAV's human cell entry receptor (AAVR) contains polycystic kidney disease (PKD) domains bound by AAV. Seeking understanding of the spectrum of interactions, goat AAVGo.1 is investigated, because its host is the species most distant from human with reciprocal cross-species cell susceptibility. The structure of AAVGo.1, solved by cryo-EM to 2.9 Å resolution, is most similar to AAV5. Through ELISA (enzyme-linked immunosorbent assay) studies, it is shown that AAVGo.1 binds to human AAVR more strongly than do AAV2 or AAV5, and that it joins AAV5 in a class that binds exclusively to PKD domain 1 (PKD1), in contrast to other AAVs that interact primarily with PKD2. The AAVGo.1 cryo-EM structure of a complex with a PKD12 fragment of AAVR at 2.4 Å resolution shows PKD1 bound with minimal change in virus structure. There are only minor conformational adaptations in AAVR, but there is a near-rigid rotation of PKD1 with maximal displacement of the receptor domain by ~1 Å compared to PKD1 bound to AAV5. AAVGo.1 joins AAV5 as the second member of an emerging class of AAVs whose mode of receptor-binding is completely different from other AAVs, typified by AAV2. Adeno-associated virus (AAV) is a small ssDNA satellite parvovirus. As a recombinant vector with a protein shell encapsidating a transgene, recombinant AAV (rAAV) is a leading delivery vehicle for gene therapy, with two FDA-approved treatments and 150 clinical trials for 30 diseases. The human entry receptor AAVR has five PKD domains. To date, all serotypes, except AAV5, have interacted primarily with the second PKD domain, PKD2. Goat is the AAV host most distant from human with cross-species cell infectivity. AAVGo.1 is similar in structure to AAV5, the two forming a class with a distinct mode of receptor-binding. Within the two classes, binding interactions are mostly conserved, giving an indication of the latitude available in modulating delivery vectors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25909.map.gz emd_25909.map.gz | 100.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25909-v30.xml emd-25909-v30.xml emd-25909.xml emd-25909.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25909.png emd_25909.png | 88.3 KB | ||
| Filedesc metadata |  emd-25909.cif.gz emd-25909.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25909 http://ftp.pdbj.org/pub/emdb/structures/EMD-25909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25909 | HTTPS FTP |
-Related structure data
| Related structure data |  7ti4MC  7ti5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25909.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25909.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of AAV-Go.1 virus like particle at 2.9 Angstroms resolution. Pixel size of 0.661 Angstroms. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66109 Å | ||||||||||||||||||||||||||||||||||||
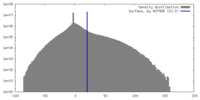
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:   Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus
| Supramolecule | Name: Adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Expressed using SF9 cells with a pfastbac LIC vector. Purified with cesium chloride ultracentrifugation. NCBI-ID: 272636 / Sci species name: Adeno-associated virus / Sci species strain: Go.1 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: Capsid, VP3 / Diameter: 250.0 Å |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Adeno-associated virus Adeno-associated virus |
| Molecular weight | Theoretical: 80.691594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFVDHPPDW LEEVGEGLRE FLGLEAGPPK PKPNQQHQDQ ARGLVLPGYN YLGPGNGLDR GEPVNRADEV AREHDISYNE QLEAGDNPY LKYNHADAEF QEKLADDTSF GGNLGKAVFQ AKKRVLEPFG LVEEGAKTAP TGKRIDDHFP KRKKARTEED S KPSTSSDA ...String: MSFVDHPPDW LEEVGEGLRE FLGLEAGPPK PKPNQQHQDQ ARGLVLPGYN YLGPGNGLDR GEPVNRADEV AREHDISYNE QLEAGDNPY LKYNHADAEF QEKLADDTSF GGNLGKAVFQ AKKRVLEPFG LVEEGAKTAP TGKRIDDHFP KRKKARTEED S KPSTSSDA EAGPSGSQQL QIPAQPASSL GADTMSAGGG GPLGDNNQGA DGVGNASGDW HCDSTWMGDR VVTKSTRTWV LP SYNNHQY REIKSGSVDG SNANAYFGYS TPWGYFDFNR FHSHWSPRDW QRLINNYWGF RPRSLRVKIF NIQVKEVTVQ DST TTIANN LTSTVQVFTD DDYQLPYVVG NGTEGCLPAF PPQVFTLPQY GYATLNRDNG DNPTERSSFF CLEYFPSKML RTGN NFEFT YSFEEVPFHC SFAPSQNLFK LANPLVDQYL YRFVSTSATG AIQFQKNLAG RYANTYKNWF PGPMGRTQGW NTSSG SSTN RVSVNNFSVS NRMNLEGASY QVNPQPNGMT NTLQGSNRYA LENTMIFNAQ NATPGTTSVY PEDNLLLTSE SETQPV NRV AYNTGGQMAT NAQNATTAPT VGTYNLQEVL PGSVWMERDV YLQGPIWAKI PETGAHFHPS PAMGGFGLKH PPPMMLI KN TPVPGNITSF SDVPVSSFIT QYSTGQVTVE MEWELKKENS KRWNPEIQYT NNYNDPQFVD FAPDGSGEYR TTRAIGTR Y LTRPL UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV Details: Two 2uL aliquots applied to grid (manual blotting between), prior to automated 3 second blot before plunging.. | ||||||||||||
| Details | Monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Average electron dose: 32.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)