[English] 日本語
 Yorodumi
Yorodumi- EMDB-25903: Cryo-electron microscopy of Adeno-associated virus serotype 4 at 2.2 A -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy of Adeno-associated virus serotype 4 at 2.2 A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAV4 / adeno-associated virus / serotype 4 / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP1/VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / Capsid Function and homology information Function and homology information | |||||||||
| Biological species |  Adeno-associated virus - 4 / Adeno-associated virus - 4 /   Adeno-associated virus Adeno-associated virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.21 Å | |||||||||
 Authors Authors | Zane GM / Silveria MA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2023 Journal: Acta Crystallogr D Struct Biol / Year: 2023Title: Cryo-EM structure of adeno-associated virus 4 at 2.2 Å resolution. Authors: Grant Zane / Mark Silveria / Nancy Meyer / Tommi White / Rui Duan / Xiaoqin Zou / Michael Chapman /  Abstract: Adeno-associated virus (AAV) is the vector of choice for several approved gene-therapy treatments and is the basis for many ongoing clinical trials. Various strains of AAV exist (referred to as ...Adeno-associated virus (AAV) is the vector of choice for several approved gene-therapy treatments and is the basis for many ongoing clinical trials. Various strains of AAV exist (referred to as serotypes), each with their own transfection characteristics. Here, a high-resolution cryo-electron microscopy structure (2.2 Å) of AAV serotype 4 (AAV4) is presented. The receptor responsible for transduction of the AAV4 clade of AAV viruses (including AAV11, AAV12 and AAVrh32.33) is unknown. Other AAVs interact with the same cell receptor, adeno-associated virus receptor (AAVR), in one of two different ways. AAV5-like viruses interact exclusively with the polycystic kidney disease-like 1 (PKD1) domain of AAVR, while most other AAVs interact primarily with the PKD2 domain. A comparison of the present AAV4 structure with prior corresponding structures of AAV5, AAV2 and AAV1 in complex with AAVR provides a foundation for understanding why the AAV4-like clade is unable to interact with either PKD1 or PKD2 of AAVR. The conformation of the AAV4 capsid in variable regions I, III, IV and V on the viral surface appears to be sufficiently different from AAV2 to ablate binding with PKD2. Differences between AAV4 and AAV5 in variable region VII appear to be sufficient to exclude binding with PKD1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25903.map.gz emd_25903.map.gz | 240.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25903-v30.xml emd-25903-v30.xml emd-25903.xml emd-25903.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25903_fsc.xml emd_25903_fsc.xml | 16 KB | Display |  FSC data file FSC data file |
| Images |  emd_25903.png emd_25903.png | 250.3 KB | ||
| Filedesc metadata |  emd-25903.cif.gz emd-25903.cif.gz | 6.8 KB | ||
| Others |  emd_25903_half_map_1.map.gz emd_25903_half_map_1.map.gz emd_25903_half_map_2.map.gz emd_25903_half_map_2.map.gz | 673.8 MB 673.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25903 http://ftp.pdbj.org/pub/emdb/structures/EMD-25903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25903 | HTTPS FTP |
-Related structure data
| Related structure data |  7thrMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25903.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25903.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.50799 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_25903_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
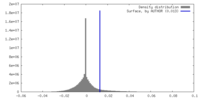
| Density Histograms |
-Half map: #2
| File | emd_25903_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
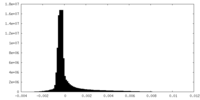
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:   Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus
| Supramolecule | Name: Adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 272636 / Sci species name: Adeno-associated virus / Sci species strain: Serotype 4 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.746 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 250.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid
| Macromolecule | Name: Capsid / type: protein_or_peptide / ID: 1 / Details: VP1 sequence of AAV4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Adeno-associated virus - 4 Adeno-associated virus - 4 |
| Molecular weight | Theoretical: 80.688023 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TTDGYLPDWL EDNLSEGVRE WWALQPGAPK PKANQQHQDN ARGLVLPGYK YLGPGNGLDK GEPVNAADAA ALEHDKAYDQ QLKAGDNPY LKYNHADAEF QQRLQGDTSF GGNLGRAVFQ AKKRVLEPLG LVEQAGETAP GKKRPLIESP QQPDSSTGIG K KGKQPAKK ...String: TTDGYLPDWL EDNLSEGVRE WWALQPGAPK PKANQQHQDN ARGLVLPGYK YLGPGNGLDK GEPVNAADAA ALEHDKAYDQ QLKAGDNPY LKYNHADAEF QQRLQGDTSF GGNLGRAVFQ AKKRVLEPLG LVEQAGETAP GKKRPLIESP QQPDSSTGIG K KGKQPAKK KLVFEDETGA GDGPPEGSTS GAMSDDSEMR AAAGGAAVEG GQGADGVGNA SGDWHCDSTW SEGHVTTTST RT WVLPTYN NHLYKRLGES LQSNTYNGFS TPWGYFDFNR FHCHFSPRDW QRLINNNWGM RPKAMRVKIF NIQVKEVTTS NGE TTVANN LTSTVQIFAD SSYELPYVMD AGQEGSLPPF PNDVFMVPQY GYCGLVTGNT SQQQTDRNAF YCLEYFPSQM LRTG NNFEI TYSFEKVPFH SMYAHSQSLD RLMNPLIDQY LWGLQSTTTG TTLNAGTATT NFTKLRPTNF SNFKKNWLPG PSIKQ QGFS KTANQNYKIP ATGSDSLIKY ETHSTLDGRW SALTPGPPMA TAGPADSKFS NSQLIFAGPK QNGNTATVPG TLIFTS EEE LAATNATDTD MWGNLPGGDQ SNSNLPTVDR LTALGAVPGM VWQNRDIYYQ GPIWAKIPHT DGHFHPSPLI GGFGLKH PP PQIFIKNTPV PANPATTFSS TPVNSFITQY STGQVSVQID WEIQKERSKR WNPEVQFTSN YGQQNSLLWA PDAAGKYT E PRAIGTRYLT HHL UniProtKB: Capsid |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 219 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.33 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: blot force: 4 time: 2 s. | |||||||||
| Details | The sample was isolated by multiple rounds of cesium chloride density centrifugation and dialyzed into a HEPES-buffered, sodium chloride solution (see Meyer et al., 2019 Bioprotocols paper). |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4809 / Average exposure time: 1.4 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 16500 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 211-734 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Link to CNS_RSRef software: https://chapman.missouri.edu/software/ |
| Output model |  PDB-7thr: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)