[English] 日本語
 Yorodumi
Yorodumi- EMDB-25837: Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ ATPase / protease / hexamer / ADP-bound state / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / ATP-dependent peptidase activity / protein catabolic process / metalloendopeptidase activity / ATP hydrolysis activity / proteolysis / zinc ion binding / ATP binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||
 Authors Authors | Liu W / Schoonen M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state. Authors: Wu Liu / Martien Schoonen / Tong Wang / Sean McSweeney / Qun Liu /  Abstract: AAA+ proteases regulate numerous physiological and cellular processes through tightly regulated proteolytic cleavage of protein substrates driven by ATP hydrolysis. FtsH is the only known family of ...AAA+ proteases regulate numerous physiological and cellular processes through tightly regulated proteolytic cleavage of protein substrates driven by ATP hydrolysis. FtsH is the only known family of membrane-anchored AAA+ proteases essential for membrane protein quality control. Although a spiral staircase rotation mechanism for substrate translocation across the FtsH pore has been proposed, the detailed conformational changes among various states have not been clear due to absence of FtsH structures in these states. We report here the cryo-EM structure for Thermotoga maritima FtsH (TmFtsH) in a fully ADP-bound symmetric state. Comparisons of the ADP-state structure with its apo-state and a substrate-engaged yeast YME1 structure show conformational changes in the ATPase domains, rather than the protease domains. A reconstruction of the full-length TmFtsH provides structural insights for the dynamic transmembrane and the periplasmic domains. Our structural analyses expand the understanding of conformational switches between different nucleotide states in ATP hydrolysis by FtsH. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25837.map.gz emd_25837.map.gz | 15.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25837-v30.xml emd-25837-v30.xml emd-25837.xml emd-25837.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
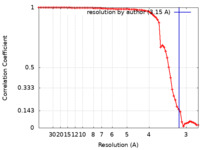
| FSC (resolution estimation) |  emd_25837_fsc.xml emd_25837_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_25837.png emd_25837.png | 177.2 KB | ||
| Filedesc metadata |  emd-25837.cif.gz emd-25837.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25837 http://ftp.pdbj.org/pub/emdb/structures/EMD-25837 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25837 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25837 | HTTPS FTP |
-Related structure data
| Related structure data |  7tdoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25837.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25837.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3684 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Hexameric FtsH in the ADP-bound state
| Entire | Name: Hexameric FtsH in the ADP-bound state |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric FtsH in the ADP-bound state
| Supramolecule | Name: Hexameric FtsH in the ADP-bound state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
-Macromolecule #1: ATP-dependent zinc metalloprotease FtsH
| Macromolecule | Name: ATP-dependent zinc metalloprotease FtsH / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
| Molecular weight | Theoretical: 68.489664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMNRSNIW NLLFTILIIV TLFWLARFFY VENSPVSKLS YTSFVQMVED ERSVVSEVVI RDDGVLRVYT KDGRVYEVDA PWAVNDSQL IEKLVSKGIK VSGERSGSSS FWINVLGTLI PTILFIVVWL FIMRSLSGRN NQAFTFTKSR ATMYKPSGNK R VTFKDVGG ...String: SNAMNRSNIW NLLFTILIIV TLFWLARFFY VENSPVSKLS YTSFVQMVED ERSVVSEVVI RDDGVLRVYT KDGRVYEVDA PWAVNDSQL IEKLVSKGIK VSGERSGSSS FWINVLGTLI PTILFIVVWL FIMRSLSGRN NQAFTFTKSR ATMYKPSGNK R VTFKDVGG AEEAIEELKE VVEFLKDPSK FNRIGARMPK GILLVGPPGT GKTLLARAVA GEANVPFFHI SGSDFVELFV GV GAARVRD LFAQAKAHAP CIVFIDEIDA VGRHRGAGLG GGHDEREQTL NQLLVEMDGF DSKEGIIVMA ATNRPDILDP ALL RPGRFD KKIVVDPPDM LGRKKILEIH TRNKPLAEDV NLEIIAKRTP GFVGADLENL VNEAALLAAR EGRDKITMKD FEEA IDRVI AGPARKSKLI SPKEKRIIAY YEAGHAVVST VVPNGEPVHR ISIIPRGYKA LGYTLHLPEE DKYLVSRNEL LDKLT ALLG GRAAEEVVFG DVTSGAANDI ERATEIARNM VCQLGMSEEL GPLAWGKEEQ EVFLGKEITR LRNYSEEVAS KIDEEV KKI VTNCYERAKE IIRKYRKQLD NIVEILLEKE TIEGDELRRI LSEEFEKVVE UniProtKB: ATP-dependent zinc metalloprotease FtsH |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)