+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
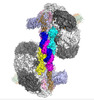
| Title | Cryo-EM structure of the Csy-AcrIF24-promoter DNA dimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Csy / Cascade / CRISPR / anti-CRISPR / AcrIF24 / HYDROLASE-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / defense response to virus / endonuclease activity / Hydrolases; Acting on ester bonds / RNA binding Similarity search - Function | |||||||||
| Biological species |  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Mukherjee IA / Chang L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2022 Journal: Nat Chem Biol / Year: 2022Title: Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor. Authors: Indranil Arun Mukherjee / Clinton Gabel / Nicholas Noinaj / Joseph Bondy-Denomy / Leifu Chang /  Abstract: Anti-CRISPR (Acr) proteins are encoded by phages to inactivate CRISPR-Cas systems of bacteria and archaea and are used to enhance the CRISPR toolbox for genome editing. Here we report the structure ...Anti-CRISPR (Acr) proteins are encoded by phages to inactivate CRISPR-Cas systems of bacteria and archaea and are used to enhance the CRISPR toolbox for genome editing. Here we report the structure and mechanism of AcrIF24, an Acr protein that inhibits the type I-F CRISPR-Cas system from Pseudomonas aeruginosa. AcrIF24 is a homodimer that associates with two copies of the surveillance complex (Csy) and prevents the hybridization between CRISPR RNA and target DNA. Furthermore, AcrIF24 functions as an anti-CRISPR-associated (Aca) protein to repress the transcription of the acrIF23-acrIF24 operon. Alone or in complex with Csy, AcrIF24 is capable of binding to the acrIF23-acrIF24 promoter DNA with nanomolar affinity. The structure of a Csy-AcrIF24-promoter DNA complex at 2.7 Å reveals the mechanism for transcriptional suppression. Our results reveal that AcrIF24 functions as an Acr-Aca fusion protein, and they extend understanding of the diverse mechanisms used by Acr proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25788.map.gz emd_25788.map.gz | 63.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25788-v30.xml emd-25788-v30.xml emd-25788.xml emd-25788.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25788.png emd_25788.png | 146.5 KB | ||
| Filedesc metadata |  emd-25788.cif.gz emd-25788.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25788 http://ftp.pdbj.org/pub/emdb/structures/EMD-25788 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25788 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25788 | HTTPS FTP |
-Validation report
| Summary document |  emd_25788_validation.pdf.gz emd_25788_validation.pdf.gz | 494.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25788_full_validation.pdf.gz emd_25788_full_validation.pdf.gz | 494.2 KB | Display | |
| Data in XML |  emd_25788_validation.xml.gz emd_25788_validation.xml.gz | 6.7 KB | Display | |
| Data in CIF |  emd_25788_validation.cif.gz emd_25788_validation.cif.gz | 7.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25788 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25788 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25788 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25788 | HTTPS FTP |
-Related structure data
| Related structure data |  7tawMC  7t3jC  7t3kC  7t3lC  7taxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25788.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25788.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.054 Å | ||||||||||||||||||||||||||||||||||||
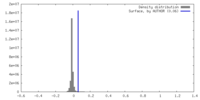
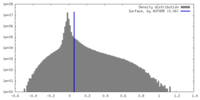
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Csy-AcrIF
| Entire | Name: Csy-AcrIF |
|---|---|
| Components |
|
-Supramolecule #1: Csy-AcrIF
| Supramolecule | Name: Csy-AcrIF / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 400 kDa/nm |
-Macromolecule #1: CRISPR-associated protein Csy1
| Macromolecule | Name: CRISPR-associated protein Csy1 / type: protein_or_peptide / ID: 1 / Details: Pseudomonas / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 49.194168 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLQPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EALQALSDNA EQAREWRQAF I GITTVKGA ...String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLQPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EALQALSDNA EQAREWRQAF I GITTVKGA PASHSLAKQL YFPLPGSGYH LLAPLFPTSL VHHVHALLRE ARFGDAAKAA REARSRQESW PHGFSEYPNL AI QKFGGTK PQNISQLNNE RRGENWLLPS LPPNWQRQNV NAPMRHSSVF EHDFGRTPEV SRLTRTLQRF LAKTVHNNLA IRQ RRAQLV AQICDEALQY AARLRELEPG WSATPGCQLH DAEQLWLDPL RAQTDETFLQ RRLRGDWPAE VGNRFANWLN RAVS SDSQI LGSPEAAQWS QELSKELTMF KEILEDERD UniProtKB: CRISPR-associated protein Csy1 |
-Macromolecule #2: CRISPR type I-F/YPEST-associated protein Csy2
| Macromolecule | Name: CRISPR type I-F/YPEST-associated protein Csy2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 36.244074 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ ...String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ RRKNQRRLTR RLLPGFALVS REALLQQHLE TLRTTLPEAT TLDALLDLCR INFEPPATSS EEEASPPDAA WQ VRDKPGW LVPIPAGYNA LSPLYLPGEV RNARDRETPL RFVENLFGLG EWLSPHRVAA LSDLLWYHHA EPDKGLYRWS TPR FVEHAI A UniProtKB: Uncharacterized protein |
-Macromolecule #3: CRISPR-associated endonuclease Cas6/Csy4
| Macromolecule | Name: CRISPR-associated endonuclease Cas6/Csy4 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 21.427504 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDHYLDIRLR PDPEFPPAQL MSVLFGKLHQ ALVAQGGDRI GVSFPDLDES RSRLGERLRI HASADDLRAL LARPWLEGLR DHLQFGEPA VVPHPTPYRQ VSRVQAKSNP ERLRRRLMRR HDLSEEEARK RIPDTVARAL DLPFVTLRSQ STGQHFRLFI R HGPLQVTA EEGGFTCYGL SKGGFVPWF UniProtKB: CRISPR-associated endonuclease Cas6/Csy4 |
-Macromolecule #4: CRISPR type I-F/YPEST-associated protein Csy3
| Macromolecule | Name: CRISPR type I-F/YPEST-associated protein Csy3 / type: protein_or_peptide / ID: 4 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 39.778594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSSHHHHHH ENLYFQSNAS KPILSTASVL AFERKLDPSD ALMSAGAWAQ RDASQEWPAV TVREKSVRGT ISNRLKTKDR DPAKLDASI QSPNLQTVDV ANLPSDADTL KVRFTLRVLG GAGTPSACND AAYRDKLLQT VATYVNDQGF AELARRYAHN L ANARFLWR ...String: MKSSHHHHHH ENLYFQSNAS KPILSTASVL AFERKLDPSD ALMSAGAWAQ RDASQEWPAV TVREKSVRGT ISNRLKTKDR DPAKLDASI QSPNLQTVDV ANLPSDADTL KVRFTLRVLG GAGTPSACND AAYRDKLLQT VATYVNDQGF AELARRYAHN L ANARFLWR NRVGAEAVEV RINHIRQGEV ARAWRFDALA IGLRDFKADA ELDALAELIA SGLSGSGHVL LEVVAFARIG DG QEVFPSQ ELILDKGDKK GQKSKTLYSV RDAAAIHSQK IGNALRTIDT WYPDEDGLGP IAVEPYGSVT SQGKAYRQPK QKL DFYTLL DNWVLRDEAP AVEQQHYVIA NLIRGGVFGE AEEK UniProtKB: CRISPR type I-F/YPEST-associated protein Csy3 |
-Macromolecule #5: AcrIF24
| Macromolecule | Name: AcrIF24 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 24.995271 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNAIHIGPFS ITPAARGLHY GGLPHHQWTL YYGPREMAIK TLPDSYTSSE VRDEFSDIIA EFVIDARHRY APDVLELVNS DGDAVLARV AVSRLPEALS GCIPDDRFPY WLLTASRPRL GLPVTLNEYT ALAVELSAPP LAWITGLLPG EVLTHDAEEW R PPTSWELR ...String: MNAIHIGPFS ITPAARGLHY GGLPHHQWTL YYGPREMAIK TLPDSYTSSE VRDEFSDIIA EFVIDARHRY APDVLELVNS DGDAVLARV AVSRLPEALS GCIPDDRFPY WLLTASRPRL GLPVTLNEYT ALAVELSAPP LAWITGLLPG EVLTHDAEEW R PPTSWELR HVVGEGSFTG VSGAAAAALL GMSATNFRKY TAGDSAANRQ KISFAAWHYL LDRLGVKRAS |
-Macromolecule #6: RNA (61-MER)
| Macromolecule | Name: RNA (61-MER) / type: rna / ID: 6 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 19.538586 KDa |
| Sequence | String: CUAAGAAAUU CACGGCGGGC UUGAUGUCCG CGUCUACCUG AUUCACUGCC GUAUAGGCAG C GENBANK: GENBANK: HQ326201.1 |
-Macromolecule #7: DNA (5'-D(P*AP*TP*AP*GP*CP*TP*CP*GP*AP*TP*TP*CP*GP*AP*GP*CP*TP*AP...
| Macromolecule | Name: DNA (5'-D(P*AP*TP*AP*GP*CP*TP*CP*GP*AP*TP*TP*CP*GP*AP*GP*CP*TP*AP*A)-3') type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 5.828798 KDa |
| Sequence | String: (DA)(DT)(DA)(DG)(DC)(DT)(DC)(DG)(DA)(DT) (DT)(DC)(DG)(DA)(DG)(DC)(DT)(DA)(DA) |
-Macromolecule #8: DNA (5'-D(P*TP*TP*AP*GP*CP*TP*CP*GP*AP*AP*TP*CP*GP*AP*GP*CP*TP*AP...
| Macromolecule | Name: DNA (5'-D(P*TP*TP*AP*GP*CP*TP*CP*GP*AP*AP*TP*CP*GP*AP*GP*CP*TP*AP*T)-3') type: dna / ID: 8 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 5.819784 KDa |
| Sequence | String: (DT)(DT)(DA)(DG)(DC)(DT)(DC)(DG)(DA)(DA) (DT)(DC)(DG)(DA)(DG)(DC)(DT)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 406243 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)