[English] 日本語
 Yorodumi
Yorodumi- EMDB-25666: Rat vesicular glutamate transporter 2 (VGLUT2) in low Cl condition -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rat vesicular glutamate transporter 2 (VGLUT2) in low Cl condition | |||||||||
 Map data Map data | sharpened map for model building | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | synaptic vesicle / neurotransmitter / glutamate transport / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-glutamate uniporter activity / sodium:phosphate symporter activity / sodium-dependent phosphate transport / phosphate ion uniporter activity / pericellular basket / Organic anion transport by SLC5/17/25 transporters / neurotransmitter uptake / hyaloid vascular plexus regression / L-glutamate import / neurotransmitter loading into synaptic vesicle ...L-glutamate uniporter activity / sodium:phosphate symporter activity / sodium-dependent phosphate transport / phosphate ion uniporter activity / pericellular basket / Organic anion transport by SLC5/17/25 transporters / neurotransmitter uptake / hyaloid vascular plexus regression / L-glutamate import / neurotransmitter loading into synaptic vesicle / L-glutamate transmembrane transporter activity / potassium:proton antiporter activity / L-glutamate transmembrane transport / phosphate ion homeostasis / neurotransmitter transmembrane transporter activity / phosphate ion transport / neural retina development / regulation of synapse structure or activity / chloride channel activity / excitatory synapse / chloride channel complex / synaptic transmission, glutamatergic / hippocampus development / synaptic vesicle / synaptic vesicle membrane / presynapse / early endosome / neuron projection / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
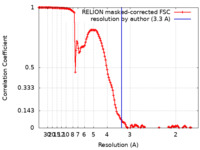
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Li F / Stroud RM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Substrate Recognition, Directional Flux, and Allostery in a Synaptic Vesicle Glutamate Transporter Authors: Li F / Stroud RM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25666.map.gz emd_25666.map.gz | 227.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25666-v30.xml emd-25666-v30.xml emd-25666.xml emd-25666.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25666_fsc.xml emd_25666_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25666.png emd_25666.png | 83.7 KB | ||
| Filedesc metadata |  emd-25666.cif.gz emd-25666.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25666 http://ftp.pdbj.org/pub/emdb/structures/EMD-25666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25666 | HTTPS FTP |
-Related structure data
| Related structure data |  7t3oMC  7t3nC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25666.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25666.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map for model building | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : complex of WT VGLUT2 with a Fab
| Entire | Name: complex of WT VGLUT2 with a Fab |
|---|---|
| Components |
|
-Supramolecule #1: complex of WT VGLUT2 with a Fab
| Supramolecule | Name: complex of WT VGLUT2 with a Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Vesicular glutamate transporter 2
| Macromolecule | Name: Vesicular glutamate transporter 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.63727 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MESVKQRILA PGKEGIKNFA GKSLGQIYRV LEKKQDNRET IELTEDGKPL EVPEKKAPLC DCTCFGLPRR YIIAIMSGLG FCISFGIRC NLGVAIVDMV NNSTIHRGGK VIKEKAKFNW DPETVGMIHG SFFWGYIITQ IPGGYIASRL AANRVFGAAI L LTSTLNML ...String: MESVKQRILA PGKEGIKNFA GKSLGQIYRV LEKKQDNRET IELTEDGKPL EVPEKKAPLC DCTCFGLPRR YIIAIMSGLG FCISFGIRC NLGVAIVDMV NNSTIHRGGK VIKEKAKFNW DPETVGMIHG SFFWGYIITQ IPGGYIASRL AANRVFGAAI L LTSTLNML IPSAARVHYG CVIFVRILQG LVEGVTYPAC HGIWSKWAPP LERSRLATTS FCGSYAGAVI AMPLAGILVQ YT GWSSVFY VYGSFGMVWY MFWLLVSYES PAKHPTITDE ERRYIEESIG ESANLLGAME KFKTPWRKFF TSMPVYAIIV ANF CRSWTF YLLLISQPAY FEEVFGFEIS KVGMLSAVPH LVMTIIVPIG GQIADFLRSK QILSTTTVRK IMNCGGFGME ATLL LVVGY SHTRGVAISF LVLAVGFSGF AISGFNVNHL DIAPRYASIL MGISNGVGTL SGMVCPIIVG AMTKNKSREE WQYVF LIAA LVHYGGVIFY ALFASGEKQP WADPEETSEE KCGFIHEDEL DEETGDITQN YINYGTTKSY GATSQENGGW PNGWEK KEE FVQESAQDAY SYKDRDDYS UniProtKB: Vesicular glutamate transporter 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 5.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)