[English] 日本語
 Yorodumi
Yorodumi- EMDB-25583: Complex of GABA-A synaptic receptor with autoimmune antibody Fab115 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of GABA-A synaptic receptor with autoimmune antibody Fab115 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | autoimmunity / encephalitis / GABA / inhibitory / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationbenzodiazepine receptor activity / GABA receptor complex / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation ...benzodiazepine receptor activity / GABA receptor complex / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation / postsynaptic specialization membrane / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / chloride channel activity / adult behavior / Signaling by ERBB4 / cochlea development / chloride channel complex / dendrite membrane / chloride transmembrane transport / cytoplasmic vesicle membrane / post-embryonic development / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / GABA-ergic synapse / dendritic spine / chemical synaptic transmission / postsynaptic membrane / postsynapse / axon / extracellular exosome / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Noviello CM / Hibbs RE / Kreye J / Teng J / Pruss H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Structural mechanisms of GABA receptor autoimmune encephalitis. Authors: Colleen M Noviello / Jakob Kreye / Jinfeng Teng / Harald Prüss / Ryan E Hibbs /   Abstract: Autoantibodies targeting neuronal membrane proteins can cause encephalitis, seizures, and severe behavioral abnormalities. While antibodies for several neuronal targets have been identified, ...Autoantibodies targeting neuronal membrane proteins can cause encephalitis, seizures, and severe behavioral abnormalities. While antibodies for several neuronal targets have been identified, structural details on how they regulate function are unknown. Here we determined cryo-electron microscopy structures of antibodies derived from an encephalitis patient bound to the γ-aminobutyric acid type A (GABA) receptor. These antibodies induced severe encephalitis by directly inhibiting GABA function, resulting in nervous-system hyperexcitability. The structures reveal mechanisms of GABA inhibition and pathology. One antibody directly competes with a neurotransmitter and locks the receptor in a resting-like state. The second antibody targets the subunit interface involved in binding benzodiazepines and antagonizes diazepam potentiation. We identify key residues in these antibodies involved in specificity and affinity and confirm structure-based hypotheses for functional effects using electrophysiology. Together these studies define mechanisms of direct functional antagonism of neurotransmission underlying autoimmune encephalitis in a human patient. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25583.map.gz emd_25583.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25583-v30.xml emd-25583-v30.xml emd-25583.xml emd-25583.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25583.png emd_25583.png | 46.2 KB | ||
| Filedesc metadata |  emd-25583.cif.gz emd-25583.cif.gz | 7.3 KB | ||
| Others |  emd_25583_additional_1.map.gz emd_25583_additional_1.map.gz emd_25583_half_map_1.map.gz emd_25583_half_map_1.map.gz emd_25583_half_map_2.map.gz emd_25583_half_map_2.map.gz | 80.7 MB 31 MB 31.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25583 http://ftp.pdbj.org/pub/emdb/structures/EMD-25583 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25583 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25583 | HTTPS FTP |
-Related structure data
| Related structure data |  7t0wMC  7t0zC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25583.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25583.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.079 Å | ||||||||||||||||||||||||||||||||||||
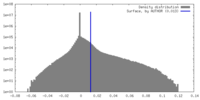
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_25583_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
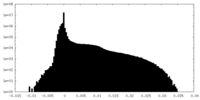
| Density Histograms |
-Half map: #1
| File | emd_25583_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25583_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between autoantibody Fab115 and the a2b2g2 GABA-A receptor
| Entire | Name: Complex between autoantibody Fab115 and the a2b2g2 GABA-A receptor |
|---|---|
| Components |
|
-Supramolecule #1: Complex between autoantibody Fab115 and the a2b2g2 GABA-A receptor
| Supramolecule | Name: Complex between autoantibody Fab115 and the a2b2g2 GABA-A receptor type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gamma-aminobutyric acid receptor subunit beta-2
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit beta-2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.40759 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVNDPSNMS LVKETVDRLL KGYDIRLRPD FGGPPVAVGM NIDIASIDMV SEVNMDYTLT MYFQQAWRDK RLSYNVIPLN LTLDNRVAD QLWVPDTYFL NDKKSFVHGV TVKNRMIRLH PDGTVLYGLR ITTTAACMMD LRRYPLDEQN CTLEIESYGY T TDDIEFYW ...String: QSVNDPSNMS LVKETVDRLL KGYDIRLRPD FGGPPVAVGM NIDIASIDMV SEVNMDYTLT MYFQQAWRDK RLSYNVIPLN LTLDNRVAD QLWVPDTYFL NDKKSFVHGV TVKNRMIRLH PDGTVLYGLR ITTTAACMMD LRRYPLDEQN CTLEIESYGY T TDDIEFYW RGDDNAVTGV TKIELPQFSI VDYKLITKKV VFSTGSYPRL SLSFKLKRNI GYFILQTYMP SILITILSWV SF WINYDAS AARVALGITT VLTMTTINTH LRETLPKIPY VKAIDMYLMG CFVFVFMALL EYALVNYIFF SQPARAAAID RWS RIFFPV VFSFFNIVYW LYYV UniProtKB: Gamma-aminobutyric acid receptor subunit beta-2, Gamma-aminobutyric acid receptor subunit beta-2 |
-Macromolecule #2: Gamma-aminobutyric acid receptor subunit alpha-1
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit alpha-1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.831836 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QPSLQDELKD NTTVFTRILD RLLDGYDNRL RPGLGERVTE VKTDIFVTSF GPVSDHDMEY TIDVFFRQSW KDERLKFKGP MTVLRLNNL MASKIWTPDT FFHNGKKSVA HNMTMPNKLL RITEDGTLLY TMRLTVRAEC PMHLEDFPMD AHACPLKFGS Y AYTRAEVV ...String: QPSLQDELKD NTTVFTRILD RLLDGYDNRL RPGLGERVTE VKTDIFVTSF GPVSDHDMEY TIDVFFRQSW KDERLKFKGP MTVLRLNNL MASKIWTPDT FFHNGKKSVA HNMTMPNKLL RITEDGTLLY TMRLTVRAEC PMHLEDFPMD AHACPLKFGS Y AYTRAEVV YEWTREPARS VVVAEDGSRL NQYDLLGQTV DSGIVQSSTG EYVVMTTHFH LKRKIGYFVI QTYLPCIMTV IL SQVSFWL NRESVPARTV FGVTTVLTMT TLSISARNSL PKVAYATAMD WFIAVCYAFV FSALIEFATV NYFTKSQPAR AAK IDRLSR IAFPLLFGIF NLVYWATYLN R UniProtKB: Gamma-aminobutyric acid receptor subunit alpha-1, Gamma-aminobutyric acid receptor subunit alpha-1 |
-Macromolecule #3: Gamma-aminobutyric acid receptor subunit gamma-2
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit gamma-2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.35466 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: WSHPQFEKGG GSGGGSGGSS AWSHPQFEKL EVLFQGPQKS DDDYEDYASN KTWVLTPKVP EGDVTVILNN LLEGYDNKLR PDIGVKPTL IHTDMYVNSI GPVNAINMEY TIDIFFAQTW YDRRLKFNST IKVLRLNSNM VGKIWIPDTF FRNSKKADAH W ITTPNRML ...String: WSHPQFEKGG GSGGGSGGSS AWSHPQFEKL EVLFQGPQKS DDDYEDYASN KTWVLTPKVP EGDVTVILNN LLEGYDNKLR PDIGVKPTL IHTDMYVNSI GPVNAINMEY TIDIFFAQTW YDRRLKFNST IKVLRLNSNM VGKIWIPDTF FRNSKKADAH W ITTPNRML RIWNDGRVLY TLRLTIDAEC QLQLHNFPMD EHSCPLEFSS YGYPREEIVY QWKRSSVEVG DTRSWRLYQF SF VGLRNTT EVVKTTSGDY VVMSVYFDLS RRMGYFTIQT YIPCTLIVVL SWVSFWINKD AVPARTSLGI TTVLTMTTLS TIA RKSLPK VSYVTAMDLF VSVCFIFVFS ALVEYGTLHY FVSSQPARAA KMDSYARIFF PTAFCLFNLV YWVSYLYL UniProtKB: Gamma-aminobutyric acid receptor subunit gamma-2, Gamma-aminobutyric acid receptor subunit gamma-2 |
-Macromolecule #4: Fab115 light chain, IgG1
| Macromolecule | Name: Fab115 light chain, IgG1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.838242 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSA SGTPGQRVTI SCSGSSSNIG SNTVSWYQQL PGTAPKLLIY STNQRPSGVP DRFSGSKSGT SASLAIGGLQ SEDEADYYC AAWDDSLKRL VVFGGGTRLT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT ...String: QSVLTQPPSA SGTPGQRVTI SCSGSSSNIG SNTVSWYQQL PGTAPKLLIY STNQRPSGVP DRFSGSKSGT SASLAIGGLQ SEDEADYYC AAWDDSLKRL VVFGGGTRLT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT PSKQSNNKYA ASSYLSLTPE QWKSHRSYSC QVTHEGSTVE KTVAPTECS |
-Macromolecule #5: Fab115 Heavy Chain, IgG1
| Macromolecule | Name: Fab115 Heavy Chain, IgG1 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.625508 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYTFI SYDINWVRQA TGQGLEWMGG MDPKSGNTGY AQKFQGRVTM TTNTAISTAY MELSSLRSE DTAVYYCVRG EQSYDRTGYS DWFDPWGQGT LVTVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS ...String: QVQLVQSGAE VKKPGASVKV SCKASGYTFI SYDINWVRQA TGQGLEWMGG MDPKSGNTGY AQKFQGRVTM TTNTAISTAY MELSSLRSE DTAVYYCVRG EQSYDRTGYS DWFDPWGQGT LVTVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPKSC DKTHDYKDDD DK HHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)