[English] 日本語
 Yorodumi
Yorodumi- EMDB-24823: Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, open-prot... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, open-protein/linear-DNA conformation | |||||||||
 Map data Map data | LocSpiral-sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNase / complex / ribonucleoprotein / genome editor / HYDROLASE-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / 3'-5' exonuclease activity / DNA endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / DNA binding / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Streptococcus pyogenes serotype M1 (bacteria) / synthetic construct (others) / Streptococcus pyogenes serotype M1 (bacteria) / synthetic construct (others) /  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) | |||||||||
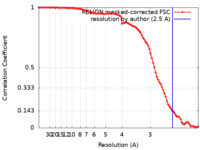
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Cofsky JC / Soczek KM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: CRISPR-Cas9 bends and twists DNA to read its sequence. Authors: Joshua C Cofsky / Katarzyna M Soczek / Gavin J Knott / Eva Nogales / Jennifer A Doudna /   Abstract: In bacterial defense and genome editing applications, the CRISPR-associated protein Cas9 searches millions of DNA base pairs to locate a 20-nucleotide, guide RNA-complementary target sequence that ...In bacterial defense and genome editing applications, the CRISPR-associated protein Cas9 searches millions of DNA base pairs to locate a 20-nucleotide, guide RNA-complementary target sequence that abuts a protospacer-adjacent motif (PAM). Target capture requires Cas9 to unwind DNA at candidate sequences using an unknown ATP-independent mechanism. Here we show that Cas9 sharply bends and undertwists DNA on PAM binding, thereby flipping DNA nucleotides out of the duplex and toward the guide RNA for sequence interrogation. Cryogenic-electron microscopy (cryo-EM) structures of Cas9-RNA-DNA complexes trapped at different states of the interrogation pathway, together with solution conformational probing, reveal that global protein rearrangement accompanies formation of an unstacked DNA hinge. Bend-induced base flipping explains how Cas9 'reads' snippets of DNA to locate target sites within a vast excess of nontarget DNA, a process crucial to both bacterial antiviral immunity and genome editing. This mechanism establishes a physical solution to the problem of complementarity-guided DNA search and shows how interrogation speed and local DNA geometry may influence genome editing efficiency. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24823.map.gz emd_24823.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24823-v30.xml emd-24823-v30.xml emd-24823.xml emd-24823.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24823_fsc.xml emd_24823_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24823.png emd_24823.png | 106.3 KB | ||
| Masks |  emd_24823_msk_1.map emd_24823_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24823.cif.gz emd-24823.cif.gz | 7.2 KB | ||
| Others |  emd_24823_additional_1.map.gz emd_24823_additional_1.map.gz emd_24823_additional_2.map.gz emd_24823_additional_2.map.gz emd_24823_half_map_1.map.gz emd_24823_half_map_1.map.gz emd_24823_half_map_2.map.gz emd_24823_half_map_2.map.gz | 30.7 MB 4.1 MB 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24823 http://ftp.pdbj.org/pub/emdb/structures/EMD-24823 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24823 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24823 | HTTPS FTP |
-Validation report
| Summary document |  emd_24823_validation.pdf.gz emd_24823_validation.pdf.gz | 908.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24823_full_validation.pdf.gz emd_24823_full_validation.pdf.gz | 907.8 KB | Display | |
| Data in XML |  emd_24823_validation.xml.gz emd_24823_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_24823_validation.cif.gz emd_24823_validation.cif.gz | 21.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24823 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24823 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24823 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24823 | HTTPS FTP |
-Related structure data
| Related structure data |  7s3hMC  7s36C  7s37C  7s38C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24823.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24823.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocSpiral-sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
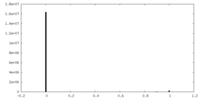
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_24823_msk_1.map emd_24823_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
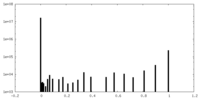
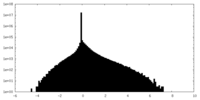
| Density Histograms |
-Additional map: unsharpened map from cryoSPARC
| File | emd_24823_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map from cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
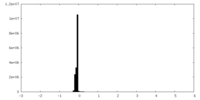
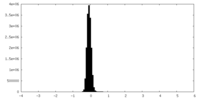
| Density Histograms |
-Additional map: post-processed map from RELION
| File | emd_24823_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post-processed map from RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
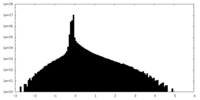
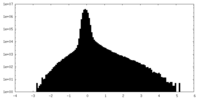
| Density Histograms |
-Half map: half map A from cryoSPARC
| File | emd_24823_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A from cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
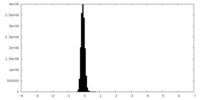
| Density Histograms |
-Half map: half map B from cryoSPARC
| File | emd_24823_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B from cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cas9 bound to sgRNA and DNA with 0bp complementarity between RNA ...
| Entire | Name: Cas9 bound to sgRNA and DNA with 0bp complementarity between RNA and DNA |
|---|---|
| Components |
|
-Supramolecule #1: Cas9 bound to sgRNA and DNA with 0bp complementarity between RNA ...
| Supramolecule | Name: Cas9 bound to sgRNA and DNA with 0bp complementarity between RNA and DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes serotype M1 (bacteria) Streptococcus pyogenes serotype M1 (bacteria) |
-Macromolecule #1: Non-target DNA strand
| Macromolecule | Name: Non-target DNA strand / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.314044 KDa |
| Sequence | String: (DG)(DA)(DC)(DG)(DC)(DA)(DT)(DA)(DA)(DA) (DG)(DA)(DT)(DG)(DA)(DG)(DA)(DC)(DA)(DA) (DT)(DG)(DG)(DC)(DG)(DA)(DT)(DT)(DA) (DC) |
-Macromolecule #4: Target DNA strand
| Macromolecule | Name: Target DNA strand / type: dna / ID: 4 Details: The 5'-most deoxycytidine is actually N4-cystamine 2'-deoxycytidine Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.190992 KDa |
| Sequence | String: (DG)(DT)(DA)(DA)(DT)(2YR)(DG)(DC)(DC)(DA) (DT)(DT)(DG)(DT)(DC)(DT)(DC)(DA)(DT) (DC)(DT)(DT)(DT)(DA)(DT)(DG)(DC)(DG)(DT) (DC) |
-Macromolecule #2: CRISPR-associated endonuclease Cas9/Csn1
| Macromolecule | Name: CRISPR-associated endonuclease Cas9/Csn1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes serotype M1 (bacteria) Streptococcus pyogenes serotype M1 (bacteria) |
| Molecular weight | Theoretical: 158.974125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMDKKYSI GLDIGTNSVG WAVITDEYKV PSKKFKVLGN TDRHSIKKNL IGALLFDSGE TAEATRLKRT ARRRYTRRKN RICYLQEIF SNEMAKVDDS FFHRLEESFL VEEDKKHERH PIFGNIVDEV AYHEKYPTIY HLRKKLVDST DKADLRLIYL A LAHMIKFR ...String: SNAMDKKYSI GLDIGTNSVG WAVITDEYKV PSKKFKVLGN TDRHSIKKNL IGALLFDSGE TAEATRLKRT ARRRYTRRKN RICYLQEIF SNEMAKVDDS FFHRLEESFL VEEDKKHERH PIFGNIVDEV AYHEKYPTIY HLRKKLVDST DKADLRLIYL A LAHMIKFR GHFLIEGDLN PDNSDVDKLF IQLVQTYNQL FEENPINASG VDAKAILSAR LSKSRRLENL IAQLPGEKKN GL FGNLIAL SLGLTPNFKS NFDLAEDAKL QLSKDTYDDD LDNLLAQIGD QYADLFLAAK NLSDAILLSD ILRVNTEITK APL SASMIK RYDEHHQDLT LLKALVRQQL PEKYKEIFFD QSKNGYAGYI DGGASQEEFY KFIKPILEKM DGTEELLVKL NRED LLRKQ RTFDNGSIPH QIHLGELHAI LRRQEDFYPF LKDNREKIEK ILTFRIPYYV GPLARGNSRF AWMTRKSEET ITPWN FEEV VDKGASAQSF IERMTNFDKN LPNEKVLPKH SLLYEYFTVY NELTKVKYVT EGMRKPAFLS GEQKKAIVDL LFKTNR KVT VKQLKEDYFK KIECFDSVEI SGVEDRFNAS LGTYHDLLKI IKDKDFLDNE ENEDILEDIV LTLTLFEDRE MIEERLK TY AHLFDDKVMK QLKRRRYTGW GRLSRKLING IRDKQSGKTI LDFLKSDGFA NRNFMQLIHD DSLTFKEDIQ KAQVSGQG D SLHEHIANLA GSPAIKKGIL QTVKVVDELV KVMGRHKPEN IVIEMARENQ TTQKGQKNSR ERMKRIEEGI KELGSQILK EHPVENTQLQ NEKLYLYYLQ NGRDMYVDQE LDINRLSDYD VDHIVPQSFL KDDSIDNKVL TRSDKNRGKS DNVPSEEVVK KMKNYWRQL LNAKLITQRK FDNLTKAERG GLSELDKAGF IKRQLVETRQ ITKHVAQILD SRMNTKYDEN DKLIREVKVI T LKSKLVSD FRKDFQFYKV REINNYHHAH DAYLNAVVGT ALIKKYPKLE SEFVYGDYKV YDVRKMIAKS EQEIGKATAK YF FYSNIMN FFKTEITLAN GEIRKRPLIE TNGETGEIVW DKGRDFATVR KVLSMPQVNI VKKTEVQTGG FSKESILPKR NSD KLIARK KDWDPKKYGG FDSPTVAYSV LVVAKVEKGK SKKLKSVKEL LGITIMERSS FEKNPIDFLE AKGYKEVKKD LIIK LPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY EKLKGSPEDN EQKQLFVEQH KHYLDEIIEQ ISEFS KRVI LADANLDKVL SAYNKHRDKP IREQAENIIH LFTLTNLGAP AAFKYFDTTI DRKRYCSTKE VLDATLIHQS ITGLYE TRI DLSQLGGD UniProtKB: CRISPR-associated endonuclease Cas9/Csn1 |
-Macromolecule #3: Single-guide RNA
| Macromolecule | Name: Single-guide RNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 32.787355 KDa |
| Sequence | String: GGCUGCGUAU UUCUACUCUG UUGUUUUAGA GCUAGAAAUA GCAAGUUAAA AUAAGGCUAG UCCGUUAUCA ACUUGAAAAA GUGGCACCG AGUCGGUGCU UCG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Details: 25mA | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)