[English] 日本語
 Yorodumi
Yorodumi- EMDB-24424: Cryo-EM structure of Thermus thermophilus reiterative transcripti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24424 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Thermus thermophilus reiterative transcription complex with 11nt oligo-G RNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Thermus thermophilus / transcription initiation / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationsigma factor activity / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / DNA-templated transcription / magnesium ion binding / DNA binding ...sigma factor activity / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) / synthetic construct (others) Thermus thermophilus HB8 (bacteria) / synthetic construct (others) | |||||||||
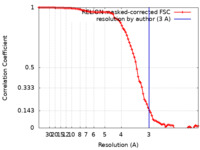
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Liu Y / Ebright RH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structural and mechanistic basis of reiterative transcription initiation. Authors: Yu Liu / Libing Yu / Chirangini Pukhrambam / Jared T Winkelman / Emre Firlar / Jason T Kaelber / Yu Zhang / Bryce E Nickels / Richard H Ebright /   Abstract: Reiterative transcription initiation, observed at promoters that contain homopolymeric sequences at the transcription start site, generates RNA products having 5' sequences noncomplementary to the ...Reiterative transcription initiation, observed at promoters that contain homopolymeric sequences at the transcription start site, generates RNA products having 5' sequences noncomplementary to the DNA template. Here, using crystallography and cryoelectron microscopy to define structures, protein-DNA photocrosslinking to map positions of RNAP leading and trailing edges relative to DNA, and single-molecule DNA nanomanipulation to assess RNA polymerase (RNAP)-dependent DNA unwinding, we show that RNA extension in reiterative transcription initiation 1) occurs without DNA scrunching; 2) involves a short, 2- to 3-bp, RNA-DNA hybrid; and 3) generates RNA that exits RNAP through the portal by which scrunched nontemplate-strand DNA exits RNAP in standard transcription initiation. The results establish that, whereas RNA extension in standard transcription initiation proceeds through a scrunching mechanism, RNA extension in reiterative transcription initiation proceeds through a slippage mechanism, with slipping of RNA relative to DNA within a short RNA-DNA hybrid, and with extrusion of RNA from RNAP through an alternative RNA exit. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24424.map.gz emd_24424.map.gz | 48.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24424-v30.xml emd-24424-v30.xml emd-24424.xml emd-24424.xml | 26.2 KB 26.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24424_fsc.xml emd_24424_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24424.png emd_24424.png | 119.3 KB | ||
| Filedesc metadata |  emd-24424.cif.gz emd-24424.cif.gz | 8.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24424 http://ftp.pdbj.org/pub/emdb/structures/EMD-24424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24424 | HTTPS FTP |
-Related structure data
| Related structure data |  7rdqMC  7mlbC  7mliC  7mljC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24424.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24424.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.038 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : reiterative transcription initiation complex with 11nt oligo-G RNA
+Supramolecule #1: reiterative transcription initiation complex with 11nt oligo-G RNA
+Macromolecule #1: DNA-directed RNA polymerase subunit alpha
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: DNA-directed RNA polymerase subunit beta'
+Macromolecule #4: DNA-directed RNA polymerase subunit omega
+Macromolecule #5: RNA polymerase sigma factor SigA
+Macromolecule #6: DNA (31-MER) nontemplate strand
+Macromolecule #7: DNA (31-MER) template strand
+Macromolecule #8: RNA (5'-R(P*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3')
+Macromolecule #9: MAGNESIUM ION
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.69 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.027 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-30 / Number grids imaged: 1 / Number real images: 3424 / Average exposure time: 0.2 sec. / Average electron dose: 0.994 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -2.8000000000000003 µm / Nominal defocus min: -1.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-7rdq: |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: B / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Output model |  PDB-7rdq: |
-Atomic model buiding 3
| Initial model | PDB ID: Chain - Chain ID: C / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Output model |  PDB-7rdq: |
-Atomic model buiding 4
| Initial model | PDB ID: Chain - Chain ID: D / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Output model |  PDB-7rdq: |
-Atomic model buiding 5
| Initial model | PDB ID: Chain - Chain ID: E / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Output model |  PDB-7rdq: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)