+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23719 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EBOV GP bound to rEBOV-442 and rEBOV-515 Fabs | |||||||||
 Map data Map data | Mucin deleted Ebola virus GP (Makona variant) in complex with rEBOV-442 and rEBOV-515 Fabs. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ebolavirus / EBOV / antibody / antibody therapeutic / mAbs / filovirus / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationclathrin-dependent endocytosis of virus by host cell / symbiont-mediated-mediated suppression of host tetherin activity / entry receptor-mediated virion attachment to host cell / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / extracellular region / membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
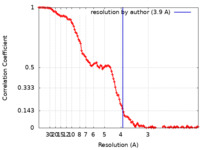
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Murin CD / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Pan-ebolavirus protective therapy by two multifunctional human antibodies. Authors: Pavlo Gilchuk / Charles D Murin / Robert W Cross / Philipp A Ilinykh / Kai Huang / Natalia Kuzmina / Viktoriya Borisevich / Krystle N Agans / Joan B Geisbert / Seth J Zost / Rachel S Nargi / ...Authors: Pavlo Gilchuk / Charles D Murin / Robert W Cross / Philipp A Ilinykh / Kai Huang / Natalia Kuzmina / Viktoriya Borisevich / Krystle N Agans / Joan B Geisbert / Seth J Zost / Rachel S Nargi / Rachel E Sutton / Naveenchandra Suryadevara / Robin G Bombardi / Robert H Carnahan / Alexander Bukreyev / Thomas W Geisbert / Andrew B Ward / James E Crowe /  Abstract: Ebolaviruses cause a severe and often fatal illness with the potential for global spread. Monoclonal antibody-based treatments that have become available recently have a narrow therapeutic spectrum ...Ebolaviruses cause a severe and often fatal illness with the potential for global spread. Monoclonal antibody-based treatments that have become available recently have a narrow therapeutic spectrum and are ineffective against ebolaviruses other than Ebola virus (EBOV), including medically important Bundibugyo (BDBV) and Sudan (SUDV) viruses. Here, we report the development of a therapeutic cocktail comprising two broadly neutralizing human antibodies, rEBOV-515 and rEBOV-442, that recognize non-overlapping sites on the ebolavirus glycoprotein (GP). Antibodies in the cocktail exhibited synergistic neutralizing activity, resisted viral escape, and possessed differing requirements for their Fc-regions for optimal in vivo activities. The cocktail protected non-human primates from ebolavirus disease caused by EBOV, BDBV, or SUDV with high therapeutic effectiveness. High-resolution structures of the cocktail antibodies in complex with GP revealed the molecular determinants for neutralization breadth and potency. This study provides advanced preclinical data to support clinical development of this cocktail for pan-ebolavirus therapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23719.map.gz emd_23719.map.gz | 7.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23719-v30.xml emd-23719-v30.xml emd-23719.xml emd-23719.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23719_fsc.xml emd_23719_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_23719.png emd_23719.png | 111.6 KB | ||
| Filedesc metadata |  emd-23719.cif.gz emd-23719.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23719 http://ftp.pdbj.org/pub/emdb/structures/EMD-23719 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23719 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23719 | HTTPS FTP |
-Validation report
| Summary document |  emd_23719_validation.pdf.gz emd_23719_validation.pdf.gz | 385.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23719_full_validation.pdf.gz emd_23719_full_validation.pdf.gz | 384.8 KB | Display | |
| Data in XML |  emd_23719_validation.xml.gz emd_23719_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_23719_validation.cif.gz emd_23719_validation.cif.gz | 16.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23719 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23719 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23719 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23719 | HTTPS FTP |
-Related structure data
| Related structure data |  7m8lMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23719.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23719.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mucin deleted Ebola virus GP (Makona variant) in complex with rEBOV-442 and rEBOV-515 Fabs. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
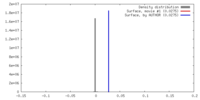
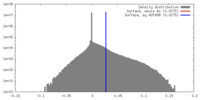
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of mucin deleted Ebola virus GP (Makona variant) bound to...
| Entire | Name: Complex of mucin deleted Ebola virus GP (Makona variant) bound to rEBOV-442 and rEBOV-515 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: Complex of mucin deleted Ebola virus GP (Makona variant) bound to...
| Supramolecule | Name: Complex of mucin deleted Ebola virus GP (Makona variant) bound to rEBOV-442 and rEBOV-515 Fabs type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Virion spike glycoprotein 1
| Macromolecule | Name: Virion spike glycoprotein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.978613 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARSIPLG VIHNSTLQVS DVDKLVCRDK LSSTNQLRSV GLNLEGNGVA TDVPSVTKR WGFRSGVPPK VVNYEAGEWA ENCYNLEIKK PDGSECLPAA PDGIRGFPRC RYVHKVSGTG PCAGDFAFHK E GAFFLYDR ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARSIPLG VIHNSTLQVS DVDKLVCRDK LSSTNQLRSV GLNLEGNGVA TDVPSVTKR WGFRSGVPPK VVNYEAGEWA ENCYNLEIKK PDGSECLPAA PDGIRGFPRC RYVHKVSGTG PCAGDFAFHK E GAFFLYDR LASTVIYRGT TFAEGVVAFL ILPQAKKDFF SSHPLREPVN ATEDPSSGYY STTIRYQATG FGTNETEYLF EV DNLTYVQ LESRFTPQFL LQLNETIYAS GKRSNTTGKL IWKVNPEIDT TIGEWAFWET KKNLTRKIRS EELSFT UniProtKB: Envelope glycoprotein |
-Macromolecule #2: rEBOV-442 Fab heavy chain
| Macromolecule | Name: rEBOV-442 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.179199 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKPGGSLRL SCTGSGGTGF TFKYAGMSWV RQAPGKGPEW IGRIKSRIDG GTTDYAAPVK DRFIVSRDDS RNTLYLQMN SLKTEDTAVY YCATGSGKGP SASFGESYYY YDFINVWGKG TTVTVSSAST KGPSVFPLAP SSKSTSGGTA A LGCLVKDY ...String: EVQLVESGGG LVKPGGSLRL SCTGSGGTGF TFKYAGMSWV RQAPGKGPEW IGRIKSRIDG GTTDYAAPVK DRFIVSRDDS RNTLYLQMN SLKTEDTAVY YCATGSGKGP SASFGESYYY YDFINVWGKG TTVTVSSAST KGPSVFPLAP SSKSTSGGTA A LGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS C |
-Macromolecule #3: rEBOV-442 Fab light chain
| Macromolecule | Name: rEBOV-442 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.715314 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ESVLTQSPGT LSLSPGERAT LSCRASQSIS RKYLAWYQQK PGQAPRLLIY GSSSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC HQYESSPWTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: ESVLTQSPGT LSLSPGERAT LSCRASQSIS RKYLAWYQQK PGQAPRLLIY GSSSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC HQYESSPWTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLRSPVTK SFNRGEC |
-Macromolecule #4: Virion spike glycoprotein 2
| Macromolecule | Name: Virion spike glycoprotein 2 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.158592 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NNNTHHQDTG EESASSGKLG LITNTIAGVA GLITGGRRTR REVIVNAQPK CNPNLHYWTT QDEGAAIGLA WIPYFGPAAE GIYTEGLMH NQDGLICGLR QLANETTQAL QLFLRATTEL RTFSILNRKA IDFLLQRWGG TCHILGPDCC IEPHDWTKNI T DKIDQIIH ...String: NNNTHHQDTG EESASSGKLG LITNTIAGVA GLITGGRRTR REVIVNAQPK CNPNLHYWTT QDEGAAIGLA WIPYFGPAAE GIYTEGLMH NQDGLICGLR QLANETTQAL QLFLRATTEL RTFSILNRKA IDFLLQRWGG TCHILGPDCC IEPHDWTKNI T DKIDQIIH DDDDKAGWSH PQFEKGGGSG GGSGGGSWSH PQFEK UniProtKB: Envelope glycoprotein |
-Macromolecule #5: rEBOV-515 Fab heavy chain
| Macromolecule | Name: rEBOV-515 Fab heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.047846 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLHESGPG LVKPSQTLSL TCTVSGGSIN SAGYYWTWIR QHPGKGLEWI GYIDYTGRTY YNPSLESRVI ISIDTSKNHF SLRLTSVSA ADTAVYYCAR ESSWVSELGR DNWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL ...String: QVQLHESGPG LVKPSQTLSL TCTVSGGSIN SAGYYWTWIR QHPGKGLEWI GYIDYTGRTY YNPSLESRVI ISIDTSKNHF SLRLTSVSA ADTAVYYCAR ESSWVSELGR DNWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSC |
-Macromolecule #6: rEBOV-515 Fab light chain
| Macromolecule | Name: rEBOV-515 Fab light chain / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.528092 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVMTQSPAT LPVSPGEGAT LSCRASQSVF TNLAWYQQKP GQAPRLLIYD ASTRATGIPA RFSGSGSGTE FTLTISSLQS EDFAVYYCQ QYNNWPRTYG QGTRVEVKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: EIVMTQSPAT LPVSPGEGAT LSCRASQSVF TNLAWYQQKP GQAPRLLIYD ASTRATGIPA RFSGSGSGTE FTLTISSLQS EDFAVYYCQ QYNNWPRTYG QGTRVEVKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLRSPVTKS FNRGEC |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)