[English] 日本語
 Yorodumi
Yorodumi- EMDB-23560: 5-fold sub-particle reconstruction from Trichomonas vaginalis vir... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23560 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 5-fold sub-particle reconstruction from Trichomonas vaginalis virus 2 capsid | |||||||||||||||||||||||||||||||||
 Map data Map data | TVV viral Capsid sub-particle (C5) | |||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | Capsid / Viral Protein | |||||||||||||||||||||||||||||||||
| Function / homology | Capsid protein Function and homology information Function and homology information | |||||||||||||||||||||||||||||||||
| Biological species |  Trichomonas vaginalis virus 2 Trichomonas vaginalis virus 2 | |||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Zhou ZH / Stevens AW | |||||||||||||||||||||||||||||||||
| Funding support |  United States, 10 items United States, 10 items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: mBio / Year: 2021 Journal: mBio / Year: 2021Title: Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2. Authors: Alexander Stevens / Katherine Muratore / Yanxiang Cui / Patricia J Johnson / Z Hong Zhou /  Abstract: , the causative pathogen for the most common nonviral sexually transmitted infection worldwide, is itself frequently infected with one or more of the four types of small double-stranded RNA (dsRNA) ..., the causative pathogen for the most common nonviral sexually transmitted infection worldwide, is itself frequently infected with one or more of the four types of small double-stranded RNA (dsRNA) viruses (TVV1 to 4, genus , family ). Each TVV encloses a nonsegmented genome within a single-layered capsid and replicates entirely intracellularly, like many dsRNA viruses, and unlike those in the family. Here, we have determined the structure of TVV2 by cryo-electron microscopy (cryoEM) at 3.6 Å resolution and derived an atomic model of its capsid. TVV2 has an icosahedral, T = 2*, capsid comprised of 60 copies of the icosahedral asymmetric unit (a dimer of the two capsid shell protein [CSP] conformers, CSP-A and CSP-B), typical of icosahedral dsRNA virus capsids. However, unlike the robust CSP-interlocking interactions such as the use of auxiliary "clamping" proteins among , only lateral CSP interactions are observed in TVV2, consistent with an assembly strategy optimized for TVVs' intracellular-only replication cycles within their protozoan host. The atomic model reveals both a mostly negatively charged capsid interior, which is conducive to movement of the loosely packed genome, and channels at the 5-fold vertices, which we suggest as routes of mRNA release during transcription. Structural comparison of TVV2 to the L-A virus reveals a conserved helix-rich fold within the CSP and putative guanylyltransferase domain along the capsid exterior, suggesting conserved mRNA maintenance strategies among This first atomic structure of a TVV provides a framework to guide future biochemical investigations into the interplay between and its viruses. viruses (TVVs) are double-stranded RNA (dsRNA) viruses that cohabitate in , the causative pathogen of trichomoniasis, the most common nonviral sexually transmitted disease worldwide. Featuring an unsegmented dsRNA genome encoding a single capsid shell protein (CSP), TVVs contrast with multisegmented dsRNA viruses, such as the diarrhea-causing rotavirus, whose larger genome is split into 10 dsRNA segments encoding 5 unique capsid proteins. To determine how TVVs incorporate the requisite functionalities for viral replication into their limited proteome, we derived the atomic model of TVV2, a first for TVVs. Our results reveal the intersubunit interactions driving CSP association for capsid assembly and the properties that govern organization and maintenance of the viral genome. Structural comparison between TVV2 capsids and those of distantly related dsRNA viruses indicates conserved strategies of nascent RNA release and a putative viral guanylyltransferase domain implicated in the cytoplasmic maintenance of viral messenger and genomic RNA. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23560.map.gz emd_23560.map.gz | 60.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23560-v30.xml emd-23560-v30.xml emd-23560.xml emd-23560.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23560.png emd_23560.png | 288.2 KB | ||
| Filedesc metadata |  emd-23560.cif.gz emd-23560.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23560 http://ftp.pdbj.org/pub/emdb/structures/EMD-23560 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23560 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23560 | HTTPS FTP |
-Related structure data
| Related structure data |  7lwyMC  7m12C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23560.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23560.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TVV viral Capsid sub-particle (C5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
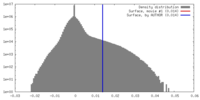
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Trichomonas vaginalis virus 2
| Entire | Name:  Trichomonas vaginalis virus 2 Trichomonas vaginalis virus 2 |
|---|---|
| Components |
|
-Supramolecule #1: Trichomonas vaginalis virus 2
| Supramolecule | Name: Trichomonas vaginalis virus 2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Virus particles isolated from lysed trichomonas vaginalis NCBI-ID: 674954 / Sci species name: Trichomonas vaginalis virus 2 / Sci species strain: 2 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Trichomonas vaginalis G3 (eukaryote) / Strain: G3 Trichomonas vaginalis G3 (eukaryote) / Strain: G3 |
| Molecular weight | Theoretical: 9.40 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 430.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Trichomonas vaginalis virus 2 Trichomonas vaginalis virus 2 |
| Molecular weight | Theoretical: 74.037117 KDa |
| Sequence | String: SNPRLTKVLD EMSKKPCVNI NEIRKMIRNF QPQFIQPRNG NRPNAQPRTV DSFEWVVRIQ STVETQLLGA TNTVPQQTLN LDISFTDDS TTITPASIPG SISMLDNSRH IPAIQSMIQN FKARYLGSLQ DTAQLQSPQY PQLLAYLFGQ LIAIKDRLDL F RPSNPLSL ...String: SNPRLTKVLD EMSKKPCVNI NEIRKMIRNF QPQFIQPRNG NRPNAQPRTV DSFEWVVRIQ STVETQLLGA TNTVPQQTLN LDISFTDDS TTITPASIPG SISMLDNSRH IPAIQSMIQN FKARYLGSLQ DTAQLQSPQY PQLLAYLFGQ LIAIKDRLDL F RPSNPLSL ADALFGFTLA QNARPRYDDH RHAKACQGPL VIPAATNSDC GPCGFVQINA NQGLTLPLGA CLFVNPETVN DQ SFQDFLW LIFATHHRMP NQMQNNWPFS LNIVSTCAAP GRQAPHAGEL TDERVRLALD TGHRILLSMF NDDEETLRYY QRK GIETMF RPCCFYTEGG LLRKATRYVS MVPLNGLYYY NGATSYVVSP IHTDAHPGIT AAIESFVDIM VLQAVFSFSG PKVV AAKVN ASQIDAAMVF GPAVAEGDGF VYDPLRPAPP LSAFYTEFIH RPAEQRIFQM AMSQIYGSHA PLIIANVINS IHNCK TKIV NNKLRATFVR RPPGAPHLKA DTAIINRFHD PELAYALGIL ADGIAPLDGS HEYNVLDELD YLFNGGDIRN CFGLNA LNT RGLGQIVHIR PKREPGKRPR RGFYTTLDGQ VHPVTQDAPL DEIYHWRDHG NLTRPYSCHI LDSQGLEFAD VSNGRSR GK ILVVVNSPLK TCAAYQGPSF APK UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: Sterile Solution was prepared fresh to prevent contamination. | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE Details: The grids were manually plunged into liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number real images: 2177 / Average exposure time: 0.2 sec. / Average electron dose: 17.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Model was manually built into EM map using Coot, then PHENIX was used for real-space refinement |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-7lwy: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)