[English] 日本語
 Yorodumi
Yorodumi- EMDB-21665: Cryo-EM map of pre-insertion mutant OCCM-DNA complex (ORC-Cdc6-Cd... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21665 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of pre-insertion mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | DNA replication / Cryo-EM / OCCM-deltaC6 / REPLICATION-DNA complex | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMCM complex loading / CDC6 association with the ORC:origin complex / Cul8-RING ubiquitin ligase complex / maintenance of rDNA / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / MCM complex binding / nuclear DNA replication / premeiotic DNA replication ...MCM complex loading / CDC6 association with the ORC:origin complex / Cul8-RING ubiquitin ligase complex / maintenance of rDNA / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / MCM complex binding / nuclear DNA replication / premeiotic DNA replication / Assembly of the ORC complex at the origin of replication / replication fork protection complex / nuclear origin of replication recognition complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / Activation of the pre-replicative complex / mitotic DNA replication / CMG complex / nuclear pre-replicative complex / cyclin-dependent protein serine/threonine kinase inhibitor activity / DNA replication preinitiation complex / Activation of ATR in response to replication stress / MCM complex / nucleosome organization / mitotic DNA replication checkpoint signaling / double-strand break repair via break-induced replication / mitotic DNA replication initiation / single-stranded DNA helicase activity / silent mating-type cassette heterochromatin formation / regulation of DNA-templated DNA replication initiation / DNA strand elongation involved in DNA replication / CDK-mediated phosphorylation and removal of Cdc6 / Orc1 removal from chromatin / nuclear replication fork / regulation of DNA replication / DNA replication origin binding / DNA replication initiation / subtelomeric heterochromatin formation / nucleosome binding / DNA helicase activity / transcription elongation by RNA polymerase II / G1/S transition of mitotic cell cycle / helicase activity / heterochromatin formation / single-stranded DNA binding / chromosome / DNA helicase / DNA replication / chromosome, telomeric region / cell division / GTPase activity / DNA damage response / chromatin binding / GTP binding / ATP hydrolysis activity / zinc ion binding / nucleoplasm / ATP binding / metal ion binding / nucleus / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.1 Å | ||||||||||||||||||
 Authors Authors | Yuan Z / Schneider S | ||||||||||||||||||
| Funding support |  United States, United States,  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6. Authors: Zuanning Yuan / Sarah Schneider / Thomas Dodd / Alberto Riera / Lin Bai / Chunli Yan / Indiana Magdalou / Ivaylo Ivanov / Bruce Stillman / Huilin Li / Christian Speck /   Abstract: DNA replication origins serve as sites of replicative helicase loading. In all eukaryotes, the six-subunit origin recognition complex (Orc1-6; ORC) recognizes the replication origin. During late M- ...DNA replication origins serve as sites of replicative helicase loading. In all eukaryotes, the six-subunit origin recognition complex (Orc1-6; ORC) recognizes the replication origin. During late M-phase of the cell-cycle, Cdc6 binds to ORC and the ORC-Cdc6 complex loads in a multistep reaction and, with the help of Cdt1, the core Mcm2-7 helicase onto DNA. A key intermediate is the ORC-Cdc6-Cdt1-Mcm2-7 (OCCM) complex in which DNA has been already inserted into the central channel of Mcm2-7. Until now, it has been unclear how the origin DNA is guided by ORC-Cdc6 and inserted into the Mcm2-7 hexamer. Here, we truncated the C-terminal winged-helix-domain (WHD) of Mcm6 to slow down the loading reaction, thereby capturing two loading intermediates prior to DNA insertion in budding yeast. In "semi-attached OCCM," the Mcm3 and Mcm7 WHDs latch onto ORC-Cdc6 while the main body of the Mcm2-7 hexamer is not connected. In "pre-insertion OCCM," the main body of Mcm2-7 docks onto ORC-Cdc6, and the origin DNA is bent and positioned adjacent to the open DNA entry gate, poised for insertion, at the Mcm2-Mcm5 interface. We used molecular simulations to reveal the dynamic transition from preloading conformers to the loaded conformers in which the loading of Mcm2-7 on DNA is complete and the DNA entry gate is fully closed. Our work provides multiple molecular insights into a key event of eukaryotic DNA replication. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21665.map.gz emd_21665.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21665-v30.xml emd-21665-v30.xml emd-21665.xml emd-21665.xml | 37.2 KB 37.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21665.png emd_21665.png | 148.2 KB | ||
| Filedesc metadata |  emd-21665.cif.gz emd-21665.cif.gz | 12.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21665 http://ftp.pdbj.org/pub/emdb/structures/EMD-21665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21665 | HTTPS FTP |
-Related structure data
| Related structure data |  6wggMC  6wgcC  6wgfC  6wgiC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21665.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21665.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
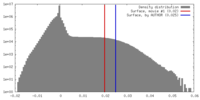
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : ORC-Cdc6-Mcm2-7-dsDNA
+Supramolecule #1: ORC-Cdc6-Mcm2-7-dsDNA
+Macromolecule #1: Cell division cycle protein CDT1
+Macromolecule #2: Cell division control protein 6
+Macromolecule #3: Origin recognition complex subunit 1
+Macromolecule #4: Origin recognition complex subunit 2
+Macromolecule #5: Origin recognition complex subunit 3
+Macromolecule #6: Origin recognition complex subunit 5
+Macromolecule #7: Origin recognition complex subunit 4
+Macromolecule #8: Origin recognition complex subunit 6
+Macromolecule #11: DNA replication licensing factor MCM2
+Macromolecule #12: DNA replication licensing factor MCM3
+Macromolecule #13: DNA replication licensing factor MCM4
+Macromolecule #14: Minichromosome maintenance protein 5
+Macromolecule #15: DNA replication licensing factor MCM6
+Macromolecule #16: DNA replication licensing factor MCM7
+Macromolecule #9: DNA (41-MER)
+Macromolecule #10: DNA (41-MER)
+Macromolecule #17: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 8.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 25126 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6wgg: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)