[English] 日本語
 Yorodumi
Yorodumi- EMDB-21247: A 2.8 Angstrom Cryo-EM Structure of a Glycoprotein B-Neutralizing... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21247 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | A 2.8 Angstrom Cryo-EM Structure of a Glycoprotein B-Neutralizing Antibody Complex Reveals a Critical Domain for Herpesvirus Fusion Initiation | |||||||||||||||
 Map data Map data | Varicella zoster virus human neutralizing antibody, 93k, in complex with native gB | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Varicella Zoster Virus / Glycoprotein B / Neutralization / Monoclonal Antibody / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / host cell endosome membrane / host cell endosome / host cell Golgi apparatus / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Human herpesvirus 3 (Varicella-zoster virus) / Human herpesvirus 3 (Varicella-zoster virus) /  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) | |||||||||||||||
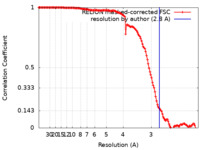
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||
 Authors Authors | Oliver SL | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: A glycoprotein B-neutralizing antibody structure at 2.8 Å uncovers a critical domain for herpesvirus fusion initiation. Authors: Stefan L Oliver / Yi Xing / Dong-Hua Chen / Soung Hun Roh / Grigore D Pintilie / David A Bushnell / Marvin H Sommer / Edward Yang / Andrea Carfi / Wah Chiu / Ann M Arvin /   Abstract: Members of the Herpesviridae, including the medically important alphaherpesvirus varicella-zoster virus (VZV), induce fusion of the virion envelope with cell membranes during entry, and between cells ...Members of the Herpesviridae, including the medically important alphaherpesvirus varicella-zoster virus (VZV), induce fusion of the virion envelope with cell membranes during entry, and between cells to form polykaryocytes in infected tissues. The conserved glycoproteins, gB, gH and gL, are the core functional proteins of the herpesvirus fusion complex. gB serves as the primary fusogen via its fusion loops, but functions for the remaining gB domains remain unexplained. As a pathway for biological discovery of domain function, our approach used structure-based analysis of the viral fusogen together with a neutralizing antibody. We report here a 2.8 Å cryogenic-electron microscopy structure of native gB recovered from VZV-infected cells, in complex with a human monoclonal antibody, 93k. This high-resolution structure guided targeted mutagenesis at the gB-93k interface, providing compelling evidence that a domain spatially distant from the gB fusion loops is critical for herpesvirus fusion, revealing a potential new target for antiviral therapies. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21247.map.gz emd_21247.map.gz | 202.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21247-v30.xml emd-21247-v30.xml emd-21247.xml emd-21247.xml | 23.9 KB 23.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21247_fsc.xml emd_21247_fsc.xml | 13.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_21247.png emd_21247.png | 170.1 KB | ||
| Filedesc metadata |  emd-21247.cif.gz emd-21247.cif.gz | 7.5 KB | ||
| Others |  emd_21247_half_map_1.map.gz emd_21247_half_map_1.map.gz emd_21247_half_map_2.map.gz emd_21247_half_map_2.map.gz | 171.1 MB 171.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21247 http://ftp.pdbj.org/pub/emdb/structures/EMD-21247 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21247 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21247 | HTTPS FTP |
-Related structure data
| Related structure data |  6vn1MC  6vlkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21247.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21247.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Varicella zoster virus human neutralizing antibody, 93k, in complex with native gB | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
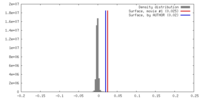
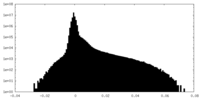
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map 1
| File | emd_21247_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
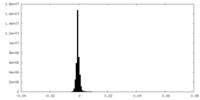
| Density Histograms |
-Half map: half map 2
| File | emd_21247_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
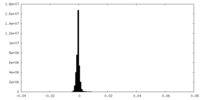
| Density Histograms |
- Sample components
Sample components
-Entire : Varicella zoster virus glycoprotein B in complex with Fab fragmen...
| Entire | Name: Varicella zoster virus glycoprotein B in complex with Fab fragments derived from human monoclonal antibody 93k |
|---|---|
| Components |
|
-Supramolecule #1: Varicella zoster virus glycoprotein B in complex with Fab fragmen...
| Supramolecule | Name: Varicella zoster virus glycoprotein B in complex with Fab fragments derived from human monoclonal antibody 93k type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 750 KDa |
-Supramolecule #2: human monoclonal antibody 93k Fab
| Supramolecule | Name: human monoclonal antibody 93k Fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Varicella zoster virus glycoprotein B
| Supramolecule | Name: Varicella zoster virus glycoprotein B / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 3 (Varicella-zoster virus) Human herpesvirus 3 (Varicella-zoster virus) |
-Macromolecule #1: Human monoclonal antibody 93k variable light chain
| Macromolecule | Name: Human monoclonal antibody 93k variable light chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.826245 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPST LSASVGDRVT ITCRASQTIS TWLAWYQQTP RKAPKLMIYK ASILENGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQ QYKSYPWTFG QGTKVEI |
-Macromolecule #2: Human monoclonal antibody 93k variable heavy chain
| Macromolecule | Name: Human monoclonal antibody 93k variable heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.658357 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGSSVKV SCKASGGTFS NFAISWVRQA PGQGLEWMGR IMPLFVTSTY AQKFQGRVTI SADASTSTAY MELSSLRSD DTAMYYCARD ITAPGAAPTP LNFYGMDVWG QGTTVTVSS |
-Macromolecule #3: Envelope glycoprotein B
| Macromolecule | Name: Envelope glycoprotein B / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Molecular weight | Theoretical: 105.50232 KDa |
| Sequence | String: MSPCGYYSKW RNRDRPEYRR NLRFRRFFSS IHPNAAAGSG FNGPGVFITS VTGVWLCFLC IFSMFVTAVV SVSPSSFYES LQVEPTQSE DITRSAHLGD GDEIREAIHK SQDAETKPTF YVCPPPTGST IVRLEPPRTC PDYHLGKNFT EGIAVVYKEN I AAYKFKAT ...String: MSPCGYYSKW RNRDRPEYRR NLRFRRFFSS IHPNAAAGSG FNGPGVFITS VTGVWLCFLC IFSMFVTAVV SVSPSSFYES LQVEPTQSE DITRSAHLGD GDEIREAIHK SQDAETKPTF YVCPPPTGST IVRLEPPRTC PDYHLGKNFT EGIAVVYKEN I AAYKFKAT VYYKDVIVST AWAGSSYTQI TNRYADRVPI PVSEITDTID KFGKCSSKAT YVRNNHKVEA FNEDKNPQDM PL IASKYNS VGSKAWHTTN DTYMVAGTPG TYRTGTSVNC IIEEVEARSI FPYDSFGLST GDIIYMSPFF GLRDGAYREH SNY AMDRFH QFEGYRQRDL DTRALLEPAA RNFLVTPHLT VGWNWKPKRT EVCSLVKWRE VEDVVRDEYA HNFRFTMKTL STTF ISETN EFNLNQIHLS QCVKEEARAI INRIYTTRYN SSHVRTGDIQ TYLARGGFVV VFQPLLSNSL ARLYLQELVR ENTNH SPQK HPTRNTRSRR SVPVELRANR TITTTSSVEF AMLQFTYDHI QEHVNEMLAR ISSSWCQLQN RERALWSGLF PINPSA LAS TILDQRVKAR ILGDVISVSN CPELGSDTRI ILQNSMRVSG STTRCYSRPL ISIVSLNGSG TVEGQLGTDN ELIMSRD LL EPCVANHKRY FLFGHHYVYY EDYRYVREIA VHDVGMISTY VDLNLTLLKD REFMPLRVYT RDELRDTGLL DYSEIQRR N QMHSLRFYDI DKVVQYDSGT AIMQGMAQFF QGLGTAGQAV GHVVLGATGA LLSTVHGFTT FLSNPFGALA VGLLVLAGL VAAFFAYRYV LKLKTSPMKA LYPLTTKGLK QLPEGMDPFA EKPNATDTPI EEIGDSQNTE PSVNSGFDPD KFREAQEMIK YMTLVSAAE RQESKARKKN KTSALLTSRL TGLALRNRRG YSRVRTENVT GV UniProtKB: ORF31 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 298 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 4 / Number real images: 11283 / Average exposure time: 12.0 sec. / Average electron dose: 7.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)