[English] 日本語
 Yorodumi
Yorodumi- EMDB-20440: Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20440 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | |||||||||
 Map data Map data | AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA 3'-5' helicase / exonuclease activity / DNA helicase activity / isomerase activity / DNA helicase / hydrolase activity / DNA repair / DNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis (bacteria) / Mycolicibacterium smegmatis (bacteria) /  Mycobacterium smegmatis (bacteria) Mycobacterium smegmatis (bacteria) | |||||||||
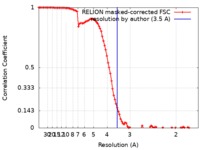
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Jia N / Unciuleac M / Shuman S / Patel DJ | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection. Authors: Ning Jia / Mihaela C Unciuleac / Chaoyou Xue / Eric C Greene / Dinshaw J Patel / Stewart Shuman /  Abstract: Mycobacterial AdnAB is a heterodimeric helicase-nuclease that initiates homologous recombination by resecting DNA double-strand breaks (DSBs). The AdnA and AdnB subunits are each composed of an N- ...Mycobacterial AdnAB is a heterodimeric helicase-nuclease that initiates homologous recombination by resecting DNA double-strand breaks (DSBs). The AdnA and AdnB subunits are each composed of an N-terminal motor domain and a C-terminal nuclease domain. Here we report cryoelectron microscopy (cryo-EM) structures of AdnAB in three functional states: in the absence of DNA and in complex with forked duplex DNAs before and after cleavage of the 5' single-strand DNA (ssDNA) tail by the AdnA nuclease. The structures reveal the path of the 5' ssDNA through the AdnA nuclease domain and the mechanism of 5' strand cleavage; the path of the 3' tracking strand through the AdnB motor and the DNA contacts that couple ATP hydrolysis to mechanical work; the position of the AdnA iron-sulfur cluster subdomain at the Y junction and its likely role in maintaining the split trajectories of the unwound 5' and 3' strands. Single-molecule DNA curtain analysis of DSB resection reveals that AdnAB is highly processive but prone to spontaneous pausing at random sites on duplex DNA. A striking property of AdnAB is that the velocity of DSB resection slows after the enzyme experiences a spontaneous pause. Our results highlight shared as well as distinctive properties of AdnAB vis-à-vis the RecBCD and AddAB clades of bacterial DSB-resecting motor nucleases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20440.map.gz emd_20440.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20440-v30.xml emd-20440-v30.xml emd-20440.xml emd-20440.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20440_fsc.xml emd_20440_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_20440.png emd_20440.png | 75.4 KB | ||
| Filedesc metadata |  emd-20440.cif.gz emd-20440.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20440 http://ftp.pdbj.org/pub/emdb/structures/EMD-20440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20440 | HTTPS FTP |
-Validation report
| Summary document |  emd_20440_validation.pdf.gz emd_20440_validation.pdf.gz | 394.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20440_full_validation.pdf.gz emd_20440_full_validation.pdf.gz | 394.3 KB | Display | |
| Data in XML |  emd_20440_validation.xml.gz emd_20440_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_20440_validation.cif.gz emd_20440_validation.cif.gz | 14.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20440 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20440 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20440 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20440 | HTTPS FTP |
-Related structure data
| Related structure data |  6ppjMC  6pprC  6ppuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20440.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20440.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8613 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AdnAB-D934A-D1014A mutant in complex with AMPPNP
| Entire | Name: AdnAB-D934A-D1014A mutant in complex with AMPPNP |
|---|---|
| Components |
|
-Supramolecule #1: AdnAB-D934A-D1014A mutant in complex with AMPPNP
| Supramolecule | Name: AdnAB-D934A-D1014A mutant in complex with AMPPNP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: UvrD/REP helicase
| Macromolecule | Name: UvrD/REP helicase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (bacteria) Mycobacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 114.735938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQVASPVVQ ARYSPVELSA ALGLFPPTDE QAAVIAAPPG PLVVIAGAGA GKTETMAARV VWLVANGFAT PSQVLGLTFT RKAAGQLLR RVRTRLARLA GAGLAPGSGA SDESATVSTY HAFAGTLLRE HGLLLPVEPD TRLLSETELW QLAYDVVCAH P GHLDTEKT ...String: MTQVASPVVQ ARYSPVELSA ALGLFPPTDE QAAVIAAPPG PLVVIAGAGA GKTETMAARV VWLVANGFAT PSQVLGLTFT RKAAGQLLR RVRTRLARLA GAGLAPGSGA SDESATVSTY HAFAGTLLRE HGLLLPVEPD TRLLSETELW QLAYDVVCAH P GHLDTEKT PAAVTAMVLR LSGALAEHLV DTDQLR(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)LLRML ATQTERTELV PLIDALHQRM RAEKVMDFGM QMAAAARLAA RFPQVGEQLR QRFRVVLLDE YQDTG HAQR IALSSLFGGG ADDGLALTAV GDPIQSIYGW RGASATNLPR FTTDFPYSDG TPAPTLELRT (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)TIRCALLNN VAAERDWVAD HLARAYHGAI GRGEAAPTAA VLVRRNADAA PMAEALTARG VPVEVV GVA GLLAVPEVAD LVAMLRLIAD PTAGSAVMRI LTGPRWRFGA RDIAALWRRA VELDDRPKGE LGTADIVAQA APDADTA CV ADAICDPGDA ERYSPAGYER IVALGRELTM LRAHLGHPLP ELVAEVRRVL GLDAEARAAR PVAAGWAGTE NLDRFSDL V SDFAGHAGAS VSALLAYLDA AVEVENGLAP AELTVSHDRV QILTVHAAKG LEWQVVAVPH LSARVFPSTT QARTWLTDA SDLPPLLRGD RATESEIGVP VLDTSDIYDR KILSDKISDH KKSLDQRRVD EERRLLYVAI TRAEDTLLLS GHHWGATESK PRGPSEFLC ELKTILEEAT AAGTPCGEIE HWAPDPAPGE TNPLRDQVVE ALWPPVASAD DHVHRGAQLV AAAMAGEVSA E ADQEGWAA DVDALLAERE RP(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)HALLG TTFHEWVQRY FHAERLFDL DDLPGAVDSD SGRAVEESLA ELQDAFVKSP WAARTPVEVE VPFDMVLGET VVRGRIDAVF AEPDGTTMVL A WKTGDPPE TPEAKEHAAV QLAVYRLAWA AMRGCPPESV RAAFHYVRSG (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)LA AAPTETAEEA DRIT UniProtKB: DNA helicase, DNA helicase, DNA helicase, DNA helicase |
-Macromolecule #2: ATP-dependent DNA helicase (UvrD/REP)
| Macromolecule | Name: ATP-dependent DNA helicase (UvrD/REP) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (bacteria) Mycobacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 110.899562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTRPAESAP QTASTLLEPG SNGVVRLLGG PGTGKSSLLV DTAVQHILAG ADPESVLLLT GSARLRTAAR AAITARLLGA GTVGVVREP LVRTVHSYAF AVLRLAAQRN GDPPPRLITS AEQDGIIREL LAGDLEDGHR SPVGWPEQLW PALTTAGFAT E LRDLMARC ...String: MTTRPAESAP QTASTLLEPG SNGVVRLLGG PGTGKSSLLV DTAVQHILAG ADPESVLLLT GSARLRTAAR AAITARLLGA GTVGVVREP LVRTVHSYAF AVLRLAAQRN GDPPPRLITS AEQDGIIREL LAGDLEDGHR SPVGWPEQLW PALTTAGFAT E LRDLMARC TERGVDPIAL QRLGRTAKRP EWLAAGRFAQ AYEQIMLLRS AVGMAAPQAT VPALGAAELV GAALEALGAD DE LLDTERN RIKLLLVDDA QHLDPQAARL VRALAAGTGL TVIAGDPDQS VFGYRGADPV LLRDDTHPAI TLTQSYRCAP EIA SAITGL GQRLPGVSDT RHWTGNPQRE GTVTVRLAAS THAEGTMIAD ALRRAHLVDG IPWSQMAVIV RSVPRVGTAL ARAL TAAGV PVQDNGTDVP VGRQPAAAAL LTVLDVTATG HLDADSAVAL LTGPIGRVDP VTLRQLRRAL RRADGSQPPR DFGDL LVDA IEREPKGLSA EHARTLRRLR AVLTAARRSD ASGADPRYTL WQAWHASGLQ RRWLAASERG GSVGAQADRD LDAVTT LFD VADQYVNRTA GASLRGLVDH VTRLGAAVAR TEPETAAEAV AVLSVHGALA GEWDFVVIAG VQEGLWPNMI PRGGVLG TQ HLVDVLDGVA DMTDRTVSTR APLVAEERRL LMAAMGRART RVMITAVDSD TGDESLLPSP FCAEISAWAT EPVAEPPL V APRVLAPSAL VGRLRAVVCA PDGAVDDDAR ACAAAQLARL AAAGVPGADP SQWHAMTSLT TEEPLWSEPG HVVTLSPST LQMLTDCPLR WLLERHGGDD GRDVRSTVGS LVHALVSEPG KTESQLVNEL EKVWDDLPYD AKWYSDNELA RHRAMLETFT RWREDTRRQ LTEVATEIPV EGIVVEPGEN TPGVRVRGRL DRLERDEAGR LVVVALKTGK SPVTKDDAQN HAQLAMYQLA V AAGLLDDG DEPGGGKLVY LGKAGAAGAT EREQDPLTPD KRAEWLETVG EAAAATAGPR FVARVNNGCA NCPVRSSCPA QA NGDRP UniProtKB: DNA helicase |
-Macromolecule #3: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 3 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #4: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 1 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Formula: Tris / Details: 20 mM Tris-HCl, pH 7.5, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.16 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)