[English] 日本語
 Yorodumi
Yorodumi- EMDB-19907: CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with GABA and puerarin | ||||||||||||
 Map data Map data | Unsharpened map from the refinement | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | GABA(A) receptor / Inhibitory synapse / GABA / Puerarin / Fat regulation / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationbenzodiazepine receptor activity / circadian sleep/wake cycle, REM sleep / reproductive behavior / GABA receptor complex / hard palate development / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity ...benzodiazepine receptor activity / circadian sleep/wake cycle, REM sleep / reproductive behavior / GABA receptor complex / hard palate development / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation / response to anesthetic / postsynaptic specialization membrane / inhibitory postsynaptic potential / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / cellular response to zinc ion / chloride channel activity / adult behavior / exploration behavior / motor behavior / roof of mouth development / Signaling by ERBB4 / cochlea development / social behavior / chloride channel complex / dendrite membrane / chloride transmembrane transport / cytoplasmic vesicle membrane / cerebellum development / post-embryonic development / learning / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / GABA-ergic synapse / memory / dendritic spine / postsynaptic membrane / postsynapse / response to xenobiotic stimulus / axon / cell surface / signal transduction / identical protein binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
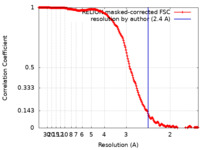
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | ||||||||||||
 Authors Authors | Kasaragod VB / Aricescu AR | ||||||||||||
| Funding support |  United Kingdom, European Union, 3 items United Kingdom, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: A brain-to-gut signal controls intestinal fat absorption. Authors: Qianqian Lyu / Wenzhi Xue / Ruixin Liu / Qinyun Ma / Vikram Babu Kasaragod / Shan Sun / Qian Li / Yanru Chen / Mingyang Yuan / Yuying Yang / Bing Zhang / Aifang Nie / Sheng Jia / Chongrong ...Authors: Qianqian Lyu / Wenzhi Xue / Ruixin Liu / Qinyun Ma / Vikram Babu Kasaragod / Shan Sun / Qian Li / Yanru Chen / Mingyang Yuan / Yuying Yang / Bing Zhang / Aifang Nie / Sheng Jia / Chongrong Shen / Po Gao / Weifang Rong / Chenxi Yu / Yufang Bi / Chunlei Zhang / Fajun Nan / Guang Ning / Zihe Rao / Xiuna Yang / Jiqiu Wang / Weiqing Wang /    Abstract: Although fat is a crucial source of energy in diets, excessive intake leads to obesity. Fat absorption in the gut is prevailingly thought to occur organ-autonomously by diffusion. Whether the ...Although fat is a crucial source of energy in diets, excessive intake leads to obesity. Fat absorption in the gut is prevailingly thought to occur organ-autonomously by diffusion. Whether the process is controlled by the brain-to-gut axis, however, remains largely unknown. Here we demonstrate that the dorsal motor nucleus of vagus (DMV) plays a key part in this process. Inactivation of DMV neurons reduces intestinal fat absorption and consequently causes weight loss, whereas activation of the DMV increases fat absorption and weight gain. Notably, the inactivation of a subpopulation of DMV neurons that project to the jejunum shortens the length of microvilli, thereby reducing fat absorption. Moreover, we identify a natural compound, puerarin, that mimics the suppression of the DMV-vagus pathway, which in turn leads to reduced fat absorption. Photoaffinity chemical methods and cryogenic electron microscopy of the structure of a GABA receptor-puerarin complex reveal that puerarin binds to an allosteric modulatory site. Notably, conditional Gabra1 knockout in the DMV largely abolishes puerarin-induced intestinal fat loss. In summary, we discover that suppression of the DMV-vagus-jejunum axis controls intestinal fat absorption by shortening the length of microvilli and illustrate the therapeutic potential of puerarin binding to GABRA1 in fat loss. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19907.map.gz emd_19907.map.gz | 257.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19907-v30.xml emd-19907-v30.xml emd-19907.xml emd-19907.xml | 30.1 KB 30.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19907_fsc.xml emd_19907_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_19907.png emd_19907.png | 129.4 KB | ||
| Masks |  emd_19907_msk_1.map emd_19907_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19907.cif.gz emd-19907.cif.gz | 8.6 KB | ||
| Others |  emd_19907_additional_1.map.gz emd_19907_additional_1.map.gz emd_19907_half_map_1.map.gz emd_19907_half_map_1.map.gz emd_19907_half_map_2.map.gz emd_19907_half_map_2.map.gz | 31.5 MB 474.5 MB 474.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19907 http://ftp.pdbj.org/pub/emdb/structures/EMD-19907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19907 | HTTPS FTP |
-Related structure data
| Related structure data |  9eqgMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19907.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19907.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from the refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
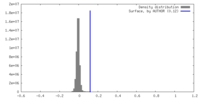
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19907_msk_1.map emd_19907_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
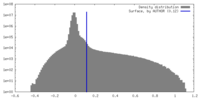
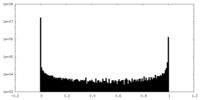
| Density Histograms |
- Sample components
Sample components
+Entire : CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R...
+Supramolecule #1: CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R...
+Macromolecule #1: Gamma-aminobutyric acid receptor subunit alpha-1
+Macromolecule #2: Gamma-aminobutyric acid receptor subunit beta-3
+Macromolecule #3: Gamma-aminobutyric acid receptor subunit gamma-2
+Macromolecule #7: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]o...
+Macromolecule #8: DECANE
+Macromolecule #9: HEXANE
+Macromolecule #10: [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tr...
+Macromolecule #11: HEXADECANE
+Macromolecule #12: PALMITIC ACID
+Macromolecule #13: CHLORIDE ION
+Macromolecule #14: GAMMA-AMINO-BUTANOIC ACID
+Macromolecule #15: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #16: Puerarin
+Macromolecule #17: 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE
+Macromolecule #18: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 15 mM Hepes pH 7.4, 100 mM NaCl | |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 287 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Temperature | Min: 84.0 K / Max: 84.0 K |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 8747 / Average exposure time: 4.19 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.5 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 96000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Real space refinement with ADP correction. |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 10 / Target criteria: FSC at 0.5 |
| Output model |  PDB-9eqg: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)