[English] 日本語
 Yorodumi
Yorodumi- EMDB-19488: Stalk-Arches-IMC at 4.33A - Refinement without symmetry of the St... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Stalk-Arches-IMC at 4.33A - Refinement without symmetry of the Stalk-Arches-IMC from the fully-assembled R388 type IV secretion. | |||||||||||||||
 Map data Map data | STALK-ARCHES-IMC _ Unsharpened | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | type IV secretion system type 4 secretion system T4SS Arches Stalk inner membrane complex IMC R388 plasmid conjugation bacterial secretion secretion secretion system protein complex VirB3 VirB4 VirB5 VirB6 VirB8 VirB10 TrwM TrwK TrwJ TrwI TrwG TrwE / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein secretion by the type IV secretion system / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
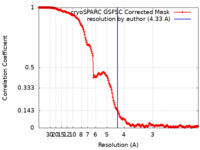
| Method | single particle reconstruction / cryo EM / Resolution: 4.33 Å | |||||||||||||||
 Authors Authors | Mace K / Waksman G | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: Cryo-EM structure of a conjugative type IV secretion system suggests a molecular switch regulating pilus biogenesis. Authors: Kévin Macé / Gabriel Waksman /   Abstract: Conjugative type IV secretion systems (T4SS) mediate bacterial conjugation, a process that enables the unidirectional exchange of genetic materials between a donor and a recipient bacterial cell. ...Conjugative type IV secretion systems (T4SS) mediate bacterial conjugation, a process that enables the unidirectional exchange of genetic materials between a donor and a recipient bacterial cell. Bacterial conjugation is the primary means by which antibiotic resistance genes spread among bacterial populations (Barlow 2009; Virolle et al, 2020). Conjugative T4SSs form pili: long extracellular filaments that connect with recipient cells. Previously, we solved the cryo-electron microscopy (cryo-EM) structure of a conjugative T4SS. In this article, based on additional data, we present a more complete T4SS cryo-EM structure than that published earlier. Novel structural features include details of the mismatch symmetry within the OMCC, the presence of a fourth VirB8 subunit in the asymmetric unit of both the arches and the inner membrane complex (IMC), and a hydrophobic VirB5 tip in the distal end of the stalk. Additionally, we provide previously undescribed structural insights into the protein VirB10 and identify a novel regulation mechanism of T4SS-mediated pilus biogenesis by this protein, that we believe is a key checkpoint for this process. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19488.map.gz emd_19488.map.gz | 251.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19488-v30.xml emd-19488-v30.xml emd-19488.xml emd-19488.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19488_fsc.xml emd_19488_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_19488.png emd_19488.png | 75.6 KB | ||
| Masks |  emd_19488_msk_1.map emd_19488_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19488.cif.gz emd-19488.cif.gz | 7 KB | ||
| Others |  emd_19488_additional_1.map.gz emd_19488_additional_1.map.gz emd_19488_half_map_1.map.gz emd_19488_half_map_1.map.gz emd_19488_half_map_2.map.gz emd_19488_half_map_2.map.gz | 455.4 MB 475.2 MB 475.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19488 http://ftp.pdbj.org/pub/emdb/structures/EMD-19488 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19488 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19488 | HTTPS FTP |
-Validation report
| Summary document |  emd_19488_validation.pdf.gz emd_19488_validation.pdf.gz | 970.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19488_full_validation.pdf.gz emd_19488_full_validation.pdf.gz | 969.8 KB | Display | |
| Data in XML |  emd_19488_validation.xml.gz emd_19488_validation.xml.gz | 26.8 KB | Display | |
| Data in CIF |  emd_19488_validation.cif.gz emd_19488_validation.cif.gz | 35.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19488 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19488 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19488 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19488 | HTTPS FTP |
-Related structure data
| Related structure data |  8rtdMC  8rt4C  8rt5C  8rt6C  8rt7C  8rt8C  8rt9C  8rtaC  8rtbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19488.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19488.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | STALK-ARCHES-IMC _ Unsharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||
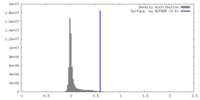
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19488_msk_1.map emd_19488_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
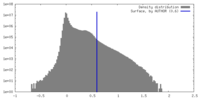
| Density Histograms |
-Additional map: STALK-ARCHES-IMC Sharpened-deepEMhancer
| File | emd_19488_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | STALK-ARCHES-IMC _ Sharpened-deepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
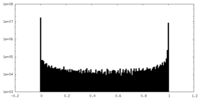
| Density Histograms |
-Half map: Half-B
| File | emd_19488_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-B | ||||||||||||
| Projections & Slices |
| ||||||||||||
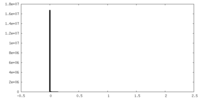
| Density Histograms |
-Half map: Half-A
| File | emd_19488_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Stalk-Arches-IMC complex from the fully-assembled R388 type IV se...
| Entire | Name: Stalk-Arches-IMC complex from the fully-assembled R388 type IV secretion system |
|---|---|
| Components |
|
-Supramolecule #1: Stalk-Arches-IMC complex from the fully-assembled R388 type IV se...
| Supramolecule | Name: Stalk-Arches-IMC complex from the fully-assembled R388 type IV secretion system type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: TrwJ protein
| Macromolecule | Name: TrwJ protein / type: protein_or_peptide / ID: 1 / Details: Sequence from conjugative plasmid R388 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.190461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKLVMTAAV AAILGAASPV MAQGIPVFDG TRALDFVQQF ARMKEQLDTA KDQLAEAQRM YEAVTGGRGL GDLMRNAQLR EYLPDDLRT VYDSANGGGY SGISGSINDI LRDERLNGSV ADMRRSIEER SRTAAATDKA VGLRAYEGAQ QRLAQIEGLM D EISRTQDQ ...String: MKKLVMTAAV AAILGAASPV MAQGIPVFDG TRALDFVQQF ARMKEQLDTA KDQLAEAQRM YEAVTGGRGL GDLMRNAQLR EYLPDDLRT VYDSANGGGY SGISGSINDI LRDERLNGSV ADMRRSIEER SRTAAATDKA VGLRAYEGAQ QRLAQIEGLM D EISRTQDQ KAIEELQARI AGEQAAIQNE TTKLQMIAQL RQAEQALISE QRRERNMRIL SSGNQGMPTI Q UniProtKB: TrwJ protein |
-Macromolecule #2: TrwG protein
| Macromolecule | Name: TrwG protein / type: protein_or_peptide / ID: 2 / Details: Sequence from conjugative plasmid R388 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.799994 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKKQPKPVK AEQLKSYYEE SRGLERDLIG EFVKSRKTAW RVATASGLFG LLGMVCGIVG FSQPAPAPLV LRVDNATGAV DVVTTLREH ESSYGEVVDT YWLNQYVLNR EAYDYNTIQM NYDTTALLSA PAVQQDYYKL FDGSNARDRV LGNKARITVR V RSIQPNGR ...String: MSKKQPKPVK AEQLKSYYEE SRGLERDLIG EFVKSRKTAW RVATASGLFG LLGMVCGIVG FSQPAPAPLV LRVDNATGAV DVVTTLREH ESSYGEVVDT YWLNQYVLNR EAYDYNTIQM NYDTTALLSA PAVQQDYYKL FDGSNARDRV LGNKARITVR V RSIQPNGR GQATVRFTTQ QHNSNGTVEA PQHQIATIGY TYIGAPMRSS DRLLNPLGFQ VTSYRADPEI LNN UniProtKB: TrwG protein |
-Macromolecule #3: TrwE protein
| Macromolecule | Name: TrwE protein / type: protein_or_peptide / ID: 3 / Details: Sequence from conjugative plasmid R388 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.443785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFGRKKGDVI DAGAELERAE QERIEGEYGA SELASERRPH TPGARTLLMV LLCVIAVVLV TLSYKAYKVR GVVEDDDAQP QQVVRQVIP GYTPRPIRPE PENVPEPPQP TTSVPAIQPA PVTQPVRPQP TGPREKTPYE LARERMLRSG LTAGSGGGED L PRPQGGDV ...String: MFGRKKGDVI DAGAELERAE QERIEGEYGA SELASERRPH TPGARTLLMV LLCVIAVVLV TLSYKAYKVR GVVEDDDAQP QQVVRQVIP GYTPRPIRPE PENVPEPPQP TTSVPAIQPA PVTQPVRPQP TGPREKTPYE LARERMLRSG LTAGSGGGED L PRPQGGDV PAGGLMGGGG GGGELAEKLQ PMRLSGSSAG RLGNRDMLIT QGTQLDCVLE TRLVTTQPGM TTCHLTRDVY ST SGRVVLL DRGSKVVGFY QGGLRQGQAR IFVQWSRIET PSGVVINLDS PGTGPLGEAG LGGWIDRHFW ERFGGAIMIS LIG DLGDWA SRQGSRQGDN SIQFSNTANG VESAAAEALR NSINIPPTLY KNQGERVNIL VARDLDFSDV YSLESIPTK UniProtKB: TrwE protein |
-Macromolecule #4: TrwI protein
| Macromolecule | Name: TrwI protein / type: protein_or_peptide / ID: 4 / Details: Sequence from conjugative plasmid R388 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.324172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAFELFTPLF NKIDQTTATY VTDISSRAIA AITPVVSVGL TLGFITYGWL IIRGAVEMPV AEFLNRCLRI GIIVSIALAG GLYQGEIAN AITTVPDELA SALLGNPTQG ASAAALVDQS AQQGFDRASE AFEEAGFFSS DGLLYGLFGI IILLATGLLA A IGGAFLLL ...String: MAFELFTPLF NKIDQTTATY VTDISSRAIA AITPVVSVGL TLGFITYGWL IIRGAVEMPV AEFLNRCLRI GIIVSIALAG GLYQGEIAN AITTVPDELA SALLGNPTQG ASAAALVDQS AQQGFDRASE AFEEAGFFSS DGLLYGLFGI IILLATGLLA A IGGAFLLL AKIALALLAG LGPLFILALI WQPTHRFFDQ WAQQVLNYGL LIVLFAAVFG LLMQIFGSYM ADLRFDGAQN VA YAIGGSV ILSIVSIVLL MQLPSIASGL AGGIGLGYMW ELRSMRSGAG AAMRGGRAMA RGARAAPGAA RGAAVGAANM AKT VATGGA GVARAAAGYF RGRKAG UniProtKB: TrwI protein |
-Macromolecule #5: TrwM protein
| Macromolecule | Name: TrwM protein / type: protein_or_peptide / ID: 5 / Details: Sequence from conjugative plasmid R388 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.292585 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKPPQQQHEA FPLFKGATRL PTIWGVPMIP LMAMVMGVAV IALTVSIWWW ALVPPLWFIM AQITKNDDKA FRIWWLWIDT KFRNRNKGF WGASSYSPAN YRKRR UniProtKB: TrwM protein |
-Macromolecule #6: Type IV secretion system protein virB4
| Macromolecule | Name: Type IV secretion system protein virB4 / type: protein_or_peptide / ID: 6 / Details: Sequence from conjugative plasmid R388 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 93.76993 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGAIESRKLL ASETPVGQFI PYSHHVTDTI ISTKNAEYLS VWKIDGRSHQ SASEADVFQW IRELNNTLRG ISSANLSLWT HIVRRRVYE YPDAEFDNVF CRQLDEKYRE SFTGYNLMVN DLYLTVVYRP VSDKVLSFFA KRERETPDQK KHRQESCIKA L EDINRTLG ...String: MGAIESRKLL ASETPVGQFI PYSHHVTDTI ISTKNAEYLS VWKIDGRSHQ SASEADVFQW IRELNNTLRG ISSANLSLWT HIVRRRVYE YPDAEFDNVF CRQLDEKYRE SFTGYNLMVN DLYLTVVYRP VSDKVLSFFA KRERETPDQK KHRQESCIKA L EDINRTLG QSFKRYGAEL LSVYEKGGHA FSAPLEFLAR LVNGEHIPMP ICRDRFSDYM AVNRPMFSKW GEVGELRSLT GL RRFGMLE IREYDDATEP GQLNVLLESD YEFVLTHSFS VLSRPAAKEY LQRHQKNLID ARDVATDQIE EIDEALNQLI SGH FVMGEH HCTLTVYGET VQQVRDNLAH ASAAMLDVAV LPKPVDLALE AGYWAQLPAN WQWRPRPAPI TSLNFLSFSP FHNF MSGKP TGNPWGPAVT ILKTVSGTPL YFNFHASKEE EDATDKRLLG NTMLIGQSSS GKTVLLGFLL AQAQKFKPTI VAFDK DRGM EISIRAMGGR YLPLKTGEPS GFNPFQLPPT HANLIFLKQF VKKLAAAGGE VTHRDEEEID QAITAMMSDS IDKSLR RLS LLLQFLPNPR SDDMDARPTV HARLVKWCEG GDYGWLFDNP TDALDLSTHQ IYGFDITEFL DNPEARTPVM MYLLYRT ES MIDGRRFMYV FDEFWKPLQD EYFEDLAKNK QKTIRKQNGI FVFATQEPSD ALESNIAKTL IQQCATYIFL ANPKADYE D YTQGFKLTDS EFELVRGLGE FSRRFLIKQG DQSALAEMNL GKFRTIVDGE TVERDFDDEL LVLSGTPDNA EIAESIIAE VGDDPAVWLP IFLDRVKAER SDV UniProtKB: Type IV secretion system protein virB4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 57.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)