+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | INTS9-INTS11-BRAT1 complex | |||||||||||||||||||||||||||
 Map data Map data | Overall map | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | Chaperone / Integrator complex / nuclear import / TRANSCRIPTION | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsnRNA 3'-end processing / snRNA processing / mitochondrion localization / integrator complex / regulation of transcription elongation by RNA polymerase II / Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters / response to ionizing radiation / RNA polymerase II transcribes snRNA genes / RNA endonuclease activity / negative regulation of transforming growth factor beta receptor signaling pathway ...snRNA 3'-end processing / snRNA processing / mitochondrion localization / integrator complex / regulation of transcription elongation by RNA polymerase II / Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters / response to ionizing radiation / RNA polymerase II transcribes snRNA genes / RNA endonuclease activity / negative regulation of transforming growth factor beta receptor signaling pathway / glucose metabolic process / cell migration / positive regulation of cell growth / blood microparticle / cell population proliferation / positive regulation of protein phosphorylation / DNA damage response / apoptotic process / nucleoplasm / membrane / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
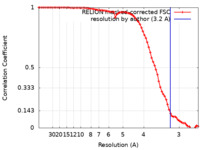
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||||||||||||||
 Authors Authors | Sabath K / Qiu C / Jonas S | |||||||||||||||||||||||||||
| Funding support |  Switzerland, European Union, 8 items Switzerland, European Union, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Assembly mechanism of Integrator's RNA cleavage module. Authors: Kevin Sabath / Chunhong Qiu / Stefanie Jonas /  Abstract: The modular Integrator complex is a transcription regulator that is essential for embryonic development. It attenuates coding gene expression via premature transcription termination and performs 3'- ...The modular Integrator complex is a transcription regulator that is essential for embryonic development. It attenuates coding gene expression via premature transcription termination and performs 3'-processing of non-coding RNAs. For both activities, Integrator requires endonuclease activity that is harbored by an RNA cleavage module consisting of INTS4-9-11. How correct assembly of Integrator modules is achieved remains unknown. Here, we show that BRAT1 and WDR73 are critical biogenesis factors for the human cleavage module. They maintain INTS9-11 inactive during maturation by physically blocking the endonuclease active site and prevent premature INTS4 association. Furthermore, BRAT1 facilitates import of INTS9-11 into the nucleus, where it is joined by INTS4. Final BRAT1 release requires locking of the mature cleavage module conformation by inositol hexaphosphate (IP). Our data explain several neurodevelopmental disorders caused by BRAT1, WDR73, and INTS11 mutations as Integrator assembly defects and reveal that IP is an essential co-factor for cleavage module maturation. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18834.map.gz emd_18834.map.gz | 3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18834-v30.xml emd-18834-v30.xml emd-18834.xml emd-18834.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18834_fsc.xml emd_18834_fsc.xml | 7.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18834.png emd_18834.png | 65 KB | ||
| Filedesc metadata |  emd-18834.cif.gz emd-18834.cif.gz | 6.8 KB | ||
| Others |  emd_18834_half_map_1.map.gz emd_18834_half_map_1.map.gz emd_18834_half_map_2.map.gz emd_18834_half_map_2.map.gz | 31.5 MB 31.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18834 http://ftp.pdbj.org/pub/emdb/structures/EMD-18834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18834 | HTTPS FTP |
-Validation report
| Summary document |  emd_18834_validation.pdf.gz emd_18834_validation.pdf.gz | 671.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18834_full_validation.pdf.gz emd_18834_full_validation.pdf.gz | 671.5 KB | Display | |
| Data in XML |  emd_18834_validation.xml.gz emd_18834_validation.xml.gz | 13.5 KB | Display | |
| Data in CIF |  emd_18834_validation.cif.gz emd_18834_validation.cif.gz | 19.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18834 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18834 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18834 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18834 | HTTPS FTP |
-Related structure data
| Related structure data |  8r23MC  8r22C  8r2dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18834.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18834.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Overall map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||
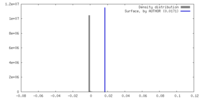
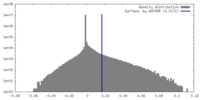
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1
| File | emd_18834_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_18834_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : INTS9-INTS11-BRAT1 complex
| Entire | Name: INTS9-INTS11-BRAT1 complex |
|---|---|
| Components |
|
-Supramolecule #1: INTS9-INTS11-BRAT1 complex
| Supramolecule | Name: INTS9-INTS11-BRAT1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: BRCA1-associated ATM activator 1
| Macromolecule | Name: BRCA1-associated ATM activator 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 88.766328 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GPSQSNMDPE CAQLLPALCA VLVDPRQPVA DDTCLEKLLD WFKTVTEGES SVVLLQEHPC LVELLSHVLK VQDLSSGVLS FSLRLAGTF AAQENCFQYL QQGELLPGLF GEPGPLGRAT WAVPTVRSGW IQGLRSLAQH PSALRFLADH GAVDTIFSLQ G DSSLFVAS ...String: GPSQSNMDPE CAQLLPALCA VLVDPRQPVA DDTCLEKLLD WFKTVTEGES SVVLLQEHPC LVELLSHVLK VQDLSSGVLS FSLRLAGTF AAQENCFQYL QQGELLPGLF GEPGPLGRAT WAVPTVRSGW IQGLRSLAQH PSALRFLADH GAVDTIFSLQ G DSSLFVAS AASQLLVHVL ALSMRGGAEG QPCLPGGDWP ACAQKIMDHV EESLCSAATP KVTQALNVLT TTFGRCQSPW TE ALWVRLS PRVACLLERD PIPAAHSFVD LLLCVARSPV FSSSDGSLWE TVARALSCLG PTHMGPLALG ILKLEHCPQA LRT QAFQVL LQPLACVLKA TVQAPGPPGL LDGTADDATT VDTLLASKSS CAGLLCRTLA HLEELQPLPQ RPSPWPQASL LGAT VTVLR LCDGSAAPAS SVGGHLCGTL AGCVRVQRAA LDFLGTLSQG TGPQELVTQA LAVLLECLES PGSSPTVLKK AFQAT LRWL LSSPKTPGCS DLGPLIPQFL RELFPVLQKR LCHPCWEVRD SALEFLTQLS RHWGGQADFR CALLASEVPQ LALQLL QDP ESYVRASAVT AMGQLSSQGL HAPTSPEHAE ARQSLFLELL HILSVDSEGF PRRAVMQVFT EWLRDGHADA AQDTEQF VA TVLQAASRDL DWEVRAQGLE LALVFLGQTL GPPRTHCPYA VALPEVAPAQ PLTEALRALC HVGLFDFAFC ALFDCDRP V AQKSCDLLLF LRDKIASYSS LREARGSPNT ASAEATLPRW RAGEQAQPPG DQEPEAVLAM LRSLDLEGLR STLAESSDH VEKSPQSLLQ DMLATGGFLQ GDEADCY UniProtKB: BRCA1-associated ATM activator 1 |
-Macromolecule #2: Integrator complex subunit 9
| Macromolecule | Name: Integrator complex subunit 9 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.891219 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKLYCLSGHP TLPCNVLKFK STTIMLDCGL DMTSTLNFLP LPLVQSPRLS NLPGWSLKDG NAFLDKELKE CSGHVFVDSV PEFCLPETE LIDLSTVDVI LISNYHCMMA LPYITEHTGF TGTVYATEPT VQIGRLLMEE LVNFIERVPK AQSASLWKNK D IQRLLPSP ...String: MKLYCLSGHP TLPCNVLKFK STTIMLDCGL DMTSTLNFLP LPLVQSPRLS NLPGWSLKDG NAFLDKELKE CSGHVFVDSV PEFCLPETE LIDLSTVDVI LISNYHCMMA LPYITEHTGF TGTVYATEPT VQIGRLLMEE LVNFIERVPK AQSASLWKNK D IQRLLPSP LKDAVEVSTW RRCYTMQEVN SALSKIQLVG YSQKIELFGA VQVTPLSSGY ALGSSNWIIQ SHYEKVSYVS GS SLLTTHP QPMDQASLKN SDVLVLTGLT QIPTANPDGM VGEFCSNLAL TVRNGGNVLV PCYPSGVIYD LLECLYQYID SAG LSSVPL YFISPVANSS LEFSQIFAEW LCHNKQSKVY LPEPPFPHAE LIQTNKLKHY PSIHGDFSND FRQPCVVFTG HPSL RFGDV VHFMELWGKS SLNTVIFTEP DFSYLEALAP YQPLAMKCIY CPIDTRLNFI QVSKLLKEVQ PLHVVCPEQY TQPPP AQSH RMDLMIDCQP PAMSYRRAEV LALPFKRRYE KIEIMPELAD SLVPMEIKPG ISLATVSAVL HTKDNKHLLQ PPPRPA QPT SGKKRKRVSD DVPDCKVLKP LLSGSIPVEQ FVQTLEKHGF SDIKVEDTAK GHIVLLQEAE TLIQIEEDST HIICDND EM LRVRLRDLVL KFLQKF UniProtKB: Integrator complex subunit 9 |
-Macromolecule #3: Integrator complex subunit 11
| Macromolecule | Name: Integrator complex subunit 11 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.997906 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GPSDPGPKRA EFMPEIRVTP LGAGQDVGRS CILVSIAGKN VMLDCGMHMG FNDDRRFPDF SYITQNGRLT DFLDCVIISH FHLDHCGAL PYFSEMVGYD GPIYMTHPTQ AICPILLEDY RKIAVDKKGE ANFFTSQMIK DCMKKVVAVH LHQTVQVDDE L EIKAYYAG ...String: GPSDPGPKRA EFMPEIRVTP LGAGQDVGRS CILVSIAGKN VMLDCGMHMG FNDDRRFPDF SYITQNGRLT DFLDCVIISH FHLDHCGAL PYFSEMVGYD GPIYMTHPTQ AICPILLEDY RKIAVDKKGE ANFFTSQMIK DCMKKVVAVH LHQTVQVDDE L EIKAYYAG HVLGAAMFQI KVGSESVVYT GDYNMTPDRH LGAAWIDKCR PNLLITESTY ATTIRDSKRC RERDFLKKVH ET VERGGKV LIPVFALGRA QELCILLETF WERMNLKVPI YFSTGLTEKA NHYYKLFIPW TNQKIRKTFV QRNMFEFKHI KAF DRAFAD NPGPMVVFAT PGMLHAGQSL QIFRKWAGNE KNMVIMPGYC VQGTVGHKIL SGQRKLEMEG RQVLEVKMQV EYMS FSAHA DAKGIMQLVG QAEPESVLLV HGEAKKMEFL KQKIEQELRV NCYMPANGET VTLPTSPSIP VGISLGLLKR EMAQG LLPE AKKPRLLHGT LIMKDSNFRL VSSEQALKEL GLAEHQLRFT CRVHLHDTRK EQETALRVYS HLKSVLKDHC VQHLPD GSV TVESVLLQAA APSEDPGTKV LLVSWTYQDE ELGSFLTSLL KKGLPQAPS UniProtKB: Integrator complex subunit 11 |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 63.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.4 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)