登録情報 データベース : EMDB / ID : EMD-18329タイトル yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate 複合体 : yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrateタンパク質・ペプチド : x 12種RNA : x 1種リガンド : x 1種 / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
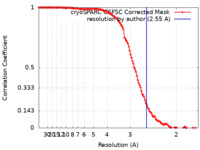
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Saccharomyces cerevisiae (パン酵母)手法 / / 解像度 : 2.55 Å Keidel A / Koegel A / Reichelt P / Kowalinski E / Schaefer IB / Conti E 資金援助 Organization Grant number 国 Max Planck Society
ジャーナル : Mol Cell / 年 : 2023タイトル : Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.著者 : Achim Keidel / Alexander Kögel / Peter Reichelt / Eva Kowalinski / Ingmar B Schäfer / Elena Conti / 要旨 : The Ski2-Ski3-Ski8 (Ski238) helicase complex directs cytoplasmic mRNAs toward the nucleolytic exosome complex for degradation. In yeast, the interaction between Ski238 and exosome requires the ... The Ski2-Ski3-Ski8 (Ski238) helicase complex directs cytoplasmic mRNAs toward the nucleolytic exosome complex for degradation. In yeast, the interaction between Ski238 and exosome requires the adaptor protein Ski7. We determined different cryo-EM structures of the Ski238 complex depicting the transition from a rigid autoinhibited closed conformation to a flexible active open conformation in which the Ski2 helicase module has detached from the rest of Ski238. The open conformation favors the interaction of the Ski3 subunit with exosome-bound Ski7, leading to the recruitment of the exosome. In the Ski238-Ski7-exosome holocomplex, the Ski2 helicase module binds the exosome cap, enabling the RNA to traverse from the helicase through the internal exosome channel to the Rrp44 exoribonuclease. Our study pinpoints how conformational changes within the Ski238 complex regulate exosome recruitment for RNA degradation. We also reveal the remarkable conservation of helicase-exosome RNA channeling mechanisms throughout eukaryotic nuclear and cytoplasmic exosome complexes. 履歴 登録 2023年8月25日 - ヘッダ(付随情報) 公開 2024年1月10日 - マップ公開 2024年1月10日 - 更新 2025年7月9日 - 現状 2025年7月9日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報
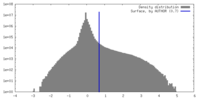
 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 データ登録者
データ登録者 ドイツ, 1件
ドイツ, 1件  引用
引用 ジャーナル: Mol Cell / 年: 2023
ジャーナル: Mol Cell / 年: 2023

 構造の表示
構造の表示 ダウンロードとリンク
ダウンロードとリンク emd_18329.map.gz
emd_18329.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-18329-v30.xml
emd-18329-v30.xml emd-18329.xml
emd-18329.xml EMDBヘッダ
EMDBヘッダ emd_18329_fsc.xml
emd_18329_fsc.xml FSCデータファイル
FSCデータファイル emd_18329.png
emd_18329.png emd_18329_msk_1.map
emd_18329_msk_1.map マスクマップ
マスクマップ emd-18329.cif.gz
emd-18329.cif.gz emd_18329_additional_1.map.gz
emd_18329_additional_1.map.gz emd_18329_additional_2.map.gz
emd_18329_additional_2.map.gz emd_18329_half_map_1.map.gz
emd_18329_half_map_1.map.gz emd_18329_half_map_2.map.gz
emd_18329_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-18329
http://ftp.pdbj.org/pub/emdb/structures/EMD-18329 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18329
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18329 emd_18329_validation.pdf.gz
emd_18329_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_18329_full_validation.pdf.gz
emd_18329_full_validation.pdf.gz emd_18329_validation.xml.gz
emd_18329_validation.xml.gz emd_18329_validation.cif.gz
emd_18329_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18329
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18329 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18329
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18329 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_18329.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_18329.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_18329_msk_1.map
emd_18329_msk_1.map 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)


















































 Trichoplusia ni (イラクサキンウワバ)
Trichoplusia ni (イラクサキンウワバ)