[English] 日本語
 Yorodumi
Yorodumi- EMDB-18328: CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state | |||||||||
 Map data Map data | Main map obtained from signal subtraction of Ski3-Nt followed by local refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helicase / RNA binding / RNA degradation / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmRNA decay by 3' to 5' exoribonuclease / protein-DNA complex assembly / Ski complex / nuclear-transcribed mRNA catabolic process, non-stop decay / nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay / reciprocal meiotic recombination / nonfunctional rRNA decay / nuclear chromosome / mRNA catabolic process / nuclear-transcribed mRNA catabolic process ...mRNA decay by 3' to 5' exoribonuclease / protein-DNA complex assembly / Ski complex / nuclear-transcribed mRNA catabolic process, non-stop decay / nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay / reciprocal meiotic recombination / nonfunctional rRNA decay / nuclear chromosome / mRNA catabolic process / nuclear-transcribed mRNA catabolic process / protein catabolic process / regulation of translation / protein-containing complex assembly / protein-macromolecule adaptor activity / defense response to virus / RNA helicase activity / RNA helicase / translation / GTPase activity / mRNA binding / protein-containing complex binding / GTP binding / ATP hydrolysis activity / DNA binding / ATP binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
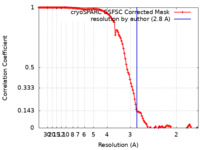
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Keidel A / Koegel A / Reichelt P / Kowalinski E / Schaefer IB / Conti E | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly. Authors: Achim Keidel / Alexander Kögel / Peter Reichelt / Eva Kowalinski / Ingmar B Schäfer / Elena Conti /   Abstract: The Ski2-Ski3-Ski8 (Ski238) helicase complex directs cytoplasmic mRNAs toward the nucleolytic exosome complex for degradation. In yeast, the interaction between Ski238 and exosome requires the ...The Ski2-Ski3-Ski8 (Ski238) helicase complex directs cytoplasmic mRNAs toward the nucleolytic exosome complex for degradation. In yeast, the interaction between Ski238 and exosome requires the adaptor protein Ski7. We determined different cryo-EM structures of the Ski238 complex depicting the transition from a rigid autoinhibited closed conformation to a flexible active open conformation in which the Ski2 helicase module has detached from the rest of Ski238. The open conformation favors the interaction of the Ski3 subunit with exosome-bound Ski7, leading to the recruitment of the exosome. In the Ski238-Ski7-exosome holocomplex, the Ski2 helicase module binds the exosome cap, enabling the RNA to traverse from the helicase through the internal exosome channel to the Rrp44 exoribonuclease. Our study pinpoints how conformational changes within the Ski238 complex regulate exosome recruitment for RNA degradation. We also reveal the remarkable conservation of helicase-exosome RNA channeling mechanisms throughout eukaryotic nuclear and cytoplasmic exosome complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18328.map.gz emd_18328.map.gz | 156.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18328-v30.xml emd-18328-v30.xml emd-18328.xml emd-18328.xml | 27.6 KB 27.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18328_fsc.xml emd_18328_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18328.png emd_18328.png | 25.9 KB | ||
| Masks |  emd_18328_msk_1.map emd_18328_msk_1.map | 166.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18328.cif.gz emd-18328.cif.gz | 8.6 KB | ||
| Others |  emd_18328_additional_1.map.gz emd_18328_additional_1.map.gz emd_18328_additional_2.map.gz emd_18328_additional_2.map.gz emd_18328_half_map_1.map.gz emd_18328_half_map_1.map.gz emd_18328_half_map_2.map.gz emd_18328_half_map_2.map.gz | 148.7 MB 156.9 MB 154.2 MB 154.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18328 http://ftp.pdbj.org/pub/emdb/structures/EMD-18328 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18328 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18328 | HTTPS FTP |
-Validation report
| Summary document |  emd_18328_validation.pdf.gz emd_18328_validation.pdf.gz | 982.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18328_full_validation.pdf.gz emd_18328_full_validation.pdf.gz | 982.3 KB | Display | |
| Data in XML |  emd_18328_validation.xml.gz emd_18328_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  emd_18328_validation.cif.gz emd_18328_validation.cif.gz | 25.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18328 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18328 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18328 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18328 | HTTPS FTP |
-Related structure data
| Related structure data |  8qcbMC  8q9tC  8qcaC  8qcfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18328.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18328.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map obtained from signal subtraction of Ski3-Nt followed by local refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18328_msk_1.map emd_18328_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: sharpened with deepEMhancer software (tightTarget)
| File | emd_18328_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened with deepEMhancer software (tightTarget) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: full map prior to signal subtraction
| File | emd_18328_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map prior to signal subtraction | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18328_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18328_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ski2delarch387 PolyU RNA
| Entire | Name: Ski2delarch387 PolyU RNA |
|---|---|
| Components |
|
-Supramolecule #1: Ski2delarch387 PolyU RNA
| Supramolecule | Name: Ski2delarch387 PolyU RNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Antiviral helicase SKI2
| Macromolecule | Name: Antiviral helicase SKI2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 146.259094 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSEGFSSSSI QELYQSLKEI TNNADVELFE DRITKLDFES TDEPKHANDI IKDRFLRPSN ALPWSLLDMV QDVPHTSSPE DCSGKLDYK ELLKVPDPIN RTSYQFKRTG LEGKISGYKE EVDLKEVANA NASNSLSITR SINHNQNSVR GSTAQLPFTP G GIPMKSVK ...String: MSEGFSSSSI QELYQSLKEI TNNADVELFE DRITKLDFES TDEPKHANDI IKDRFLRPSN ALPWSLLDMV QDVPHTSSPE DCSGKLDYK ELLKVPDPIN RTSYQFKRTG LEGKISGYKE EVDLKEVANA NASNSLSITR SINHNQNSVR GSTAQLPFTP G GIPMKSVK TDSEQNGSST MANATKLLHK DGQGLFDIPE GMNRGIKPMD SPAENEDQNG QFKELKQLNE IDNELDIRIE AN EAKLKEE EKSAKSISEE IMEEATEETT ADNADDAEID ELLPIGIDFG RTKPVSKSVP VKKEWAHVVD LNHKIENFDE LIP NPARSW PFELDTFQKE AVYHLEQGDS VFVAAHTSAG KTVVAEYAIA MAHRNMTKTI YTSPIKALSN QKFRDFKETF DDVN IGLIT GDVQINPDAN CLIMTTEILR SMLYRGADLI RDVEFVIFDE VHYVNDQDRG VVWEEVIIML PQHVKFILLS ATVPN TYEF ANWIGRTKQK NIYVISTPKR PVPLEINIWA KKELIPVINQ NSEFLEANFR KHKEILNGES AKGAPSKTDN GRGGST ARG GRGGSNTRDG RGGRGNSTRG GANRGGSRGA GAIGSNKRKF FTQDGPSKKT WPEIVNYLRK RELLPMVVFV FSKKRCE EY ADWLEGINFC NNKEKSQIHM FIEKSITRLK KEDRDLPQIL KTRSLLERGI AVHHGGLLPI VKELIEILFS KGFIKVLF A TETFAMGLNL PTRTVIFSSI RKHDGNGLRE LTPGEFTQMA GRAGRRGLDS TGTVIVMAYN SPLSIATFKE VTMGVPTRL QSQFRLTYNM ILNLLRIEAL RVEEMIKYSF SENAKETLQP EHEKQIKVLQ EELQTIEYKS CEICDNDIEK FLELMLAYKE ATVNLMQEM VKSPSILHIL KEGRLVAFRD PNDCLKLGFV FKVSLKDAVC VIMTFTKPYK LPNGEPNHLI YFPKADGYRR R NFPKFQKT DFYMEEVPVT AIEVITKRKF AAPLGKVIKK DVAALNEFNA ETNNILDGKT LKEAINIEKQ GLKIHQILLD RT NIRDEIF KLKSIKCPNL SQHIVPKFKA HVIKKKIEEL YHLMSDQNLS LLPDYEKRLA VLKDTEFIDQ NHNVLLKGRV ACE INSGYE LVLTELILDN FLGSFEPEEI VALLSVFVYE GKTREEEPPI VTPRLAKGKQ RIEEIYKKML CVFNTHQIPL TQDE AEFLD RKRFAMMNVV YEWARGLSFK EIMEMSPEAE GTVVRVITWL DEICREVKTA SIIIGNSTLH MKMSRAQELI KRDIV FAAS LYL UniProtKB: Antiviral helicase SKI2 |
-Macromolecule #2: Superkiller protein 3
| Macromolecule | Name: Superkiller protein 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 164.278484 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GPDSMSDIKQ LLKEAKQELT NRDYEETIEI SEKVLKLDPD NYFAHIFLGK ALSSLPASNN VSSNRNLERA TNHYVSAAKL VPDNLLAWK GLFLLFRTTE VVPDILSYDE YFDLCGQYAD ALLKQEQSQV ELINDIKLLK KTHPDCQKAF YQHLKPGSLM A ETIGRHLS ...String: GPDSMSDIKQ LLKEAKQELT NRDYEETIEI SEKVLKLDPD NYFAHIFLGK ALSSLPASNN VSSNRNLERA TNHYVSAAKL VPDNLLAWK GLFLLFRTTE VVPDILSYDE YFDLCGQYAD ALLKQEQSQV ELINDIKLLK KTHPDCQKAF YQHLKPGSLM A ETIGRHLS TPQDALLNLI KILSNIETTE IGKTLSQNRL KLKASDPDYQ IKLNSFSWEI IKNSEIDQLY NQLVNILADD QK RSEIENQ WLEYRIKVLK SMPLDVKKDF FTKVKEMVED MVLVNHQSLL AWQKYFEWTD YEDLDNMDAP LIIKYFKKFP KDP LAMILY SWLSSKLSKY DIKSLESANK PPEGHKKTEK ETDIKDVDET NEDEVKDRVE DEVKDRVEDE VKDQDEEAKE DEEE DLDDI EIGLLEEEVV TVLTENIVKC KNNILAHRIL CQYYLLTKEY EAALPYIKNG ISLIAYNIKD LGVHLPLTKR EFSLD LATV YTYVDAPKDH NAALKLYDNI LSGDFSNIQA KMGKGIIFIE RKNWKDAMTL LTQVHEQSPN NLEVLSELSW SKAHMG YMD EALAGLDTVI KGIKGMDLRS IDFRALNLWR QAKVYIMKHA SINDAKQENV KCAFKLLIQS IKILDTFAPG FSTLGDI YC HYYKDHLRAF KCYFKAFDLD AGDYTAAKYI TETYASKPNW QAASSIASRL IKGEKAKAEL RSNNWPFRVV GIAHLEKQ E ESDSIEWFQS ALRVDPNDVE SWVGLGQAYH ACGRIEASIK VFDKAIQLRP SHTFAQYFKA ISLCDVGEYL ESLDILEKV CQEAATEESF QIGLVEVLMR CSLDLYSQGF LLKSVSIAKD TIERIKIIIS ELKCENQQVW IYLSQVLRLF IWIESKVDTL PVESLVSIF ENSQFSGSEE IDSVDNIKID TLLDSTTDDN VSIACKFLIL ASKYSVSDQK FTDIAGTVRA SYWYNIGISE L TAFITLKE PQYRDAAIFA FKKSIQLQSN TSETWIGLGI ATMDINFRVS QHCFIKATAL EPKATNTWFN LAMLGLKKKD TE FAQQVLN KLQSLAPQDS SPWLGMALIL EEQGDIIGSS KLFAHSFILS NGRSKAAQFM YAKNVLENHI NNGDDERDIE TVE KLTTAS IALEQFFKKS PDSQFALQCA LLTLERLHHY ENANELANRL IGILEKKFEK TQDERELFNF AIIKGQFARI HLGL GNFEL SIENADLSQG IISESSDEKS MKTKISNHIC LGLSYFFLND FDQTLNQFQE LLSISKDSKH LVVLIAKVLY DVGES DTKE IALQELTEYI ATSGADLLVT LTIAAMSILD DKREDLSIIL EELKALPLSK QIIDKHKDAP YLIEEITKRL YRNDTG KQV WQRSAYFFPN NLKVWERLDK NIQRRIASNG QNKVTAEEMS KLYCESKNLR SIQRGMFLCP WNVTAVKALN ECF UniProtKB: Superkiller protein 3 |
-Macromolecule #3: Antiviral protein SKI8
| Macromolecule | Name: Antiviral protein SKI8 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.283527 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSKVFIATAN AGKAHDADIF SVSACNSFTV SCSGDGYLKV WDNKLLDNEN PKDKSYSHFV HKSGLHHVDV LQAIERDAFE LCLVATTSF SGDLLFYRIT REDETKKVIF EKLDLLDSDM KKHSFWALKW GASNDRLLSH RLVATDVKGT TYIWKFHPFA D ESNSLTLN ...String: MSKVFIATAN AGKAHDADIF SVSACNSFTV SCSGDGYLKV WDNKLLDNEN PKDKSYSHFV HKSGLHHVDV LQAIERDAFE LCLVATTSF SGDLLFYRIT REDETKKVIF EKLDLLDSDM KKHSFWALKW GASNDRLLSH RLVATDVKGT TYIWKFHPFA D ESNSLTLN WSPTLELQGT VESPMTPSQF ATSVDISERG LIATGFNNGT VQISELSTLR PLYNFESQHS MINNSNSIRS VK FSPQGSL LAIAHDSNSF GCITLYETEF GERIGSLSVP THSSQASLGE FAHSSWVMSL SFNDSGETLC SAGWDGKLRF WDV KTKERI TTLNMHCDDI EIEEDILAVD EHGDSLAEPG VFDVKFLKKG WRSGMGADLN ESLCCVCLDR SIRWFREAGG K UniProtKB: Antiviral protein SKI8 |
-Macromolecule #4: Superkiller protein 7
| Macromolecule | Name: Superkiller protein 7 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.345137 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPDSMSLLEQ LARKRIEKSK GLLSADQSHS TSKSASLLER LHKNRETKDN NAETKRKDLK TLLAKDKVKR SDFTPNQHSV SLSLKLSAL KKSNSDLEKQ GKSVTLDSKE NELPTKRKSP DDKLNLEESW KAIKEMNHYC FLKNDPCINQ TDDFAFTNFI I KDKKNSLS ...String: GPDSMSLLEQ LARKRIEKSK GLLSADQSHS TSKSASLLER LHKNRETKDN NAETKRKDLK TLLAKDKVKR SDFTPNQHSV SLSLKLSAL KKSNSDLEKQ GKSVTLDSKE NELPTKRKSP DDKLNLEESW KAIKEMNHYC FLKNDPCINQ TDDFAFTNFI I KDKKNSLS TSIPLSSQNS SFLSLKKHNN ELLGIFVPCN LPKTTRKVAI ENFNRPSPDD IIQSAQLNAF NEKLENLNIK SA GSWSHPQ FEK UniProtKB: Superkiller protein 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 7127 / Average electron dose: 77.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)