[English] 日本語
 Yorodumi
Yorodumi- EMDB-17925: Cryo-EM structure of human Elp123 in complex with 5'-deoxyadenosi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human Elp123 in complex with 5'-deoxyadenosine and methionine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Elongator / tRNA modification / acetyl-CoA hydrolysis / TRANSLATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphorylase kinase regulator activity / tRNA uridine(34) acetyltransferase activity / tRNA carboxymethyluridine synthase / elongator holoenzyme complex / tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation / tRNA wobble uridine modification / regulation of receptor signaling pathway via JAK-STAT / acetyltransferase activity / central nervous system development / transcription elongation factor complex ...phosphorylase kinase regulator activity / tRNA uridine(34) acetyltransferase activity / tRNA carboxymethyluridine synthase / elongator holoenzyme complex / tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation / tRNA wobble uridine modification / regulation of receptor signaling pathway via JAK-STAT / acetyltransferase activity / central nervous system development / transcription elongation factor complex / transcription elongation by RNA polymerase II / neuron migration / regulation of translation / HATs acetylate histones / 4 iron, 4 sulfur cluster binding / tRNA binding / positive regulation of cell migration / regulation of transcription by RNA polymerase II / protein kinase binding / nucleolus / nucleoplasm / metal ion binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
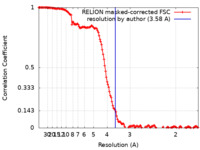
| Method | single particle reconstruction / cryo EM / Resolution: 3.58 Å | |||||||||
 Authors Authors | Abbassi N / Jaciuk M / Lin T-Y / Glatt S | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structures of the human Elongator complex at work. Authors: Nour-El-Hana Abbassi / Marcin Jaciuk / David Scherf / Pauline Böhnert / Alexander Rau / Alexander Hammermeister / Michał Rawski / Paulina Indyka / Grzegorz Wazny / Andrzej Chramiec- ...Authors: Nour-El-Hana Abbassi / Marcin Jaciuk / David Scherf / Pauline Böhnert / Alexander Rau / Alexander Hammermeister / Michał Rawski / Paulina Indyka / Grzegorz Wazny / Andrzej Chramiec-Głąbik / Dominika Dobosz / Bozena Skupien-Rabian / Urszula Jankowska / Juri Rappsilber / Raffael Schaffrath / Ting-Yu Lin / Sebastian Glatt /    Abstract: tRNA modifications affect ribosomal elongation speed and co-translational folding dynamics. The Elongator complex is responsible for introducing 5-carboxymethyl at wobble uridine bases (cmU) in ...tRNA modifications affect ribosomal elongation speed and co-translational folding dynamics. The Elongator complex is responsible for introducing 5-carboxymethyl at wobble uridine bases (cmU) in eukaryotic tRNAs. However, the structure and function of human Elongator remain poorly understood. In this study, we present a series of cryo-EM structures of human ELP123 in complex with tRNA and cofactors at four different stages of the reaction. The structures at resolutions of up to 2.9 Å together with complementary functional analyses reveal the molecular mechanism of the modification reaction. Our results show that tRNA binding exposes a universally conserved uridine at position 33 (U), which triggers acetyl-CoA hydrolysis. We identify a series of conserved residues that are crucial for the radical-based acetylation of U and profile the molecular effects of patient-derived mutations. Together, we provide the high-resolution view of human Elongator and reveal its detailed mechanism of action. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17925.map.gz emd_17925.map.gz | 15.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17925-v30.xml emd-17925-v30.xml emd-17925.xml emd-17925.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17925_fsc.xml emd_17925_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17925.png emd_17925.png | 60.4 KB | ||
| Masks |  emd_17925_msk_1.map emd_17925_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17925.cif.gz emd-17925.cif.gz | 8.4 KB | ||
| Others |  emd_17925_half_map_1.map.gz emd_17925_half_map_1.map.gz emd_17925_half_map_2.map.gz emd_17925_half_map_2.map.gz | 194.3 MB 194 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17925 http://ftp.pdbj.org/pub/emdb/structures/EMD-17925 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17925 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17925 | HTTPS FTP |
-Related structure data
| Related structure data |  8ptyMC  8ptxC  8ptzC  8pu0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17925.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17925.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17925_msk_1.map emd_17925_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17925_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17925_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human Elp123 in complex with 5'-deoxyadenosine and methionine
| Entire | Name: Human Elp123 in complex with 5'-deoxyadenosine and methionine |
|---|---|
| Components |
|
-Supramolecule #1: Human Elp123 in complex with 5'-deoxyadenosine and methionine
| Supramolecule | Name: Human Elp123 in complex with 5'-deoxyadenosine and methionine type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 610 KDa |
-Macromolecule #1: Elongator complex protein 1
| Macromolecule | Name: Elongator complex protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150.427484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRNLKLFRTL EFRDIQGPGN PQCFSLRTEQ GTVLIGSEHG LIEVDPVSRE VKNEVSLVAE GFLPEDGSGR IVGVQDLLDQ ESVCVATAS GDVILCSLST QQLECVGSVA SGISVMSWSP DQELVLLATG QQTLIMMTKD FEPILEQQIH QDDFGESKFI T VGWGRKET ...String: MRNLKLFRTL EFRDIQGPGN PQCFSLRTEQ GTVLIGSEHG LIEVDPVSRE VKNEVSLVAE GFLPEDGSGR IVGVQDLLDQ ESVCVATAS GDVILCSLST QQLECVGSVA SGISVMSWSP DQELVLLATG QQTLIMMTKD FEPILEQQIH QDDFGESKFI T VGWGRKET QFHGSEGRQA AFQMQMHESA LPWDDHRPQV TWRGDGQFFA VSVVCPETGA RKVRVWNREF ALQSTSEPVA GL GPALAWK PSGSLIASTQ DKPNQQDIVF FEKNGLLHGH FTLPFLKDEV KVNDLLWNAD SSVLAVWLED LQREESSIPK TCV QLWTVG NYHWYLKQSL SFSTCGKSKI VSLMWDPVTP YRLHVLCQGW HYLAYDWHWT TDRSVGDNSS DLSNVAVIDG NRVL VTVFR QTVVPPPMCT YQLLFPHPVN QVTFLAHPQK SNDLAVLDAS NQISVYKCGD CPSADPTVKL GAVGGSGFKV CLRTP HLEK RYKIQFENNE DQDVNPLKLG LLTWIEEDVF LAVSHSEFSP RSVIHHLTAA SSEMDEEHGQ LNVSSSAAVD GVIISL CCN SKTKSVVLQL ADGQIFKYLW ESPSLAIKPW KNSGGFPVRF PYPCTQTELA MIGEEECVLG LTDRCRFFIN DIEVASN IT SFAVYDEFLL LTTHSHTCQC FCLRDASFKT LQAGLSSNHV SHGEVLRKVE RGSRIVTVVP QDTKLVLQMP RGNLEVVH H RALVLAQIRK WLDKLMFKEA FECMRKLRIN LNLIYDHNPK VFLGNVETFI KQIDSVNHIN LFFTELKEED VTKTMYPAP VTSSVYLSRD PDGNKIDLVC DAMRAVMESI NPHKYCLSIL TSHVKKTTPE LEIVLQKVHE LQGNAPSDPD AVSAEEALKY LLHLVDVNE LYDHSLGTYD FDLVLMVAEK SQKDPKEYLP FLNTLKKMET NYQRFTIDKY LKRYEKAIGH LSKCGPEYFP E CLNLIKDK NLYNEALKLY SPSSQQYQDI SIAYGEHLMQ EHMYEPAGLM FARCGAHEKA LSAFLTCGNW KQALCVAAQL NF TKDQLVG LGRTLAGKLV EQRKHIDAAM VLEECAQDYE EAVLLLLEGA AWEEALRLVY KYNRLDIIET NVKPSILEAQ KNY MAFLDS QTATFSRHKK RLLVVRELKE QAQQAGLDDE VPHGQESDLF SETSSVVSGS EMSGKYSHSN SRISARSSKN RRKA ERKKH SLKEGSPLED LALLEALSEV VQNTENLKDE VYHILKVLFL FEFDEQGREL QKAFEDTLQL MERSLPEIWT LTYQQ NSAT PVLGPNSTAN SIMASYQQQK TSVPVLDAEL FIPPKINRRT QWKLSLLD UniProtKB: Elongator complex protein 1 |
-Macromolecule #2: Elongator complex protein 2
| Macromolecule | Name: Elongator complex protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.597766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVAPVLETSH VFCCPNRVRG VLNWSSGPRG LLAFGTSCSV VLYDPLKRVV VTNLNGHTAR VNCIQWICKQ DGSPSTELVS GGSDNQVIH WEIEDNQLLK AVHLQGHEGP VYAVHAVYQR RTSDPALCTL IVSAAADSAV RLWSKKGPEV MCLQTLNFGN G FALALCLS ...String: MVAPVLETSH VFCCPNRVRG VLNWSSGPRG LLAFGTSCSV VLYDPLKRVV VTNLNGHTAR VNCIQWICKQ DGSPSTELVS GGSDNQVIH WEIEDNQLLK AVHLQGHEGP VYAVHAVYQR RTSDPALCTL IVSAAADSAV RLWSKKGPEV MCLQTLNFGN G FALALCLS FLPNTDVPIL ACGNDDCRIH IFAQQNDQFQ KVLSLCGHED WIRGVEWAAF GRDLFLASCS QDCLIRIWKL YI KSTSLET QDDDNIRLKE NTFTIENESV KIAFAVTLET VLAGHENWVN AVHWQPVFYK DGVLQQPVRL LSASMDKTMI LWA PDEESG VWLEQVRVGE VGGNTLGFYD CQFNEDGSMI IAHAFHGALH LWKQNTVNPR EWTPEIVISG HFDGVQDLVW DPEG EFIIT VGTDQTTRLF APWKRKDQSQ VTWHEIARPQ IHGYDLKCLA MINRFQFVSG ADEKVLRVFS APRNFVENFC AITGQ SLNH VLCNQDSDLP EGATVPALGL SNKAVFQGDI ASQPSDEEEL LTSTGFEYQQ VAFQPSILTE PPTEDHLLQN TLWPEV QKL YGHGYEIFCV TCNSSKTLLA SACKAAKKEH AAIILWNTTS WKQVQNLVFH SLTVTQMAFS PNEKFLLAVS RDRTWSL WK KQDTISPEFE PVFSLFAFTN KITSVHSRII WSCDWSPDSK YFFTGSRDKK VVVWGECDST DDCIEHNIGP CSSVLDVG G AVTAVSVCPV LHPSQRYVVA VGLECGKICL YTWKKTDQVP EINDWTHCVE TSQSQSHTLA IRKLCWKNCS GKTEQKEAE GAEWLHFASC GEDHTVKIHR VNKCAL UniProtKB: Elongator complex protein 2 |
-Macromolecule #3: Elongator complex protein 3
| Macromolecule | Name: Elongator complex protein 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 65.740539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRQKRKGDLS PAELMMLTIG DVIKQLIEAH EQGKDIDLNK VKTKTAAKYG LSAQPRLVDI IAAVPPQYRK VLMPKLKAKP IRTASGIAV VAVMCKPHRC PHISFTGNIC VYCPGGPDSD FEYSTQSYTG YEPTSMRAIR ARYDPFLQTR HRIEQLKQLG H SVDKVEFI ...String: MRQKRKGDLS PAELMMLTIG DVIKQLIEAH EQGKDIDLNK VKTKTAAKYG LSAQPRLVDI IAAVPPQYRK VLMPKLKAKP IRTASGIAV VAVMCKPHRC PHISFTGNIC VYCPGGPDSD FEYSTQSYTG YEPTSMRAIR ARYDPFLQTR HRIEQLKQLG H SVDKVEFI VMGGTFMALP EEYRDYFIRN LHDALSGHTS NNIYEAVKYS ERSLTKCIGI TIETRPDYCM KRHLSDMLTY GC TRLEIGV QSVYEDVARD TNRGHTVKAV CESFHLAKDS GFKVVAHMMP DLPNVGLERD IEQFTEFFEN PAFRPDGLKL YPT LVIRGT GLYELWKSGR YKSYSPSDLV ELVARILALV PPWTRVYRVQ RDIPMPLVSS GVEHGNLREL ALARMKDLGI QCRD VRTRE VGIQEIHHKV RPYQVELVRR DYVANGGWET FLSYEDPDQD ILIGLLRLRK CSEETFRFEL GGGVSIVREL HVYGS VVPV SSRDPTKFQH QGFGMLLMEE AERIAREEHG SGKIAVISGV GTRNYYRKIG YRLQGPYMVK MLKGLEGSAW SHPQFE KGG GSGGGSGGSA WSHPQFEK UniProtKB: Elongator complex protein 3 |
-Macromolecule #4: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 4 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #5: 5'-DEOXYADENOSINE
| Macromolecule | Name: 5'-DEOXYADENOSINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: 5AD |
|---|---|
| Molecular weight | Theoretical: 251.242 Da |
| Chemical component information |  ChemComp-5AD: |
-Macromolecule #6: METHIONINE
| Macromolecule | Name: METHIONINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: MET |
|---|---|
| Molecular weight | Theoretical: 149.211 Da |
| Chemical component information |  ChemComp-MET: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR / Details: 8 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 15 s wait time, blot force 5, 5 s blot time. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Software | Name: EPU |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 5300 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: SwissModel / Chain - Initial model type: in silico model / Details: Based on PDB 6QK7 |
|---|---|
| Software | Name: UCSF ChimeraX |
| Output model |  PDB-8pty: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)