[English] 日本語
 Yorodumi
Yorodumi- EMDB-17445: Cryo-EM structure of the c-di-GMP-free FleQ-FleN master regulator... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
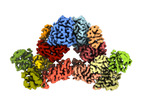
| Title | Cryo-EM structure of the c-di-GMP-free FleQ-FleN master regulator complex of P. aeruginosa | |||||||||
 Map data Map data | Sharpened cryo-EM density map of the c-di-GMP-free FleQ-FleN master regulator complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pseudomonas aeruginosa / biofilm / c-di-GMP / transcription regulation / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of cilium-dependent cell motility / regulation of bacterial-type flagellum-dependent cell motility / cyclic-di-GMP binding / positive regulation of cell-substrate adhesion / negative regulation of extracellular matrix assembly / DNA-binding transcription repressor activity / DNA-binding transcription activator activity / cis-regulatory region sequence-specific DNA binding / protein-DNA complex / cytoplasmic side of plasma membrane ...positive regulation of cilium-dependent cell motility / regulation of bacterial-type flagellum-dependent cell motility / cyclic-di-GMP binding / positive regulation of cell-substrate adhesion / negative regulation of extracellular matrix assembly / DNA-binding transcription repressor activity / DNA-binding transcription activator activity / cis-regulatory region sequence-specific DNA binding / protein-DNA complex / cytoplasmic side of plasma membrane / transcription cis-regulatory region binding / regulation of DNA-templated transcription / DNA-templated transcription / positive regulation of DNA-templated transcription / ATP hydrolysis activity / ATP binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Torres-Sanchez L / Krasteva PV | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structures of the FleQ-FleN master regulators reveal large-scale conformational switching in motility and biofilm control. Authors: Lucía Torres-Sánchez / Thibault Géry Sana / Marion Decossas / Yaser Hashem / Petya Violinova Krasteva /  Abstract: can cause a wide array of chronic and acute infections associated with its ability to rapidly switch between planktonic, biofilm, and dispersed lifestyles, each with a specific arsenal for bacterial ... can cause a wide array of chronic and acute infections associated with its ability to rapidly switch between planktonic, biofilm, and dispersed lifestyles, each with a specific arsenal for bacterial survival and virulence. At the cellular level, many of the physiological transitions are orchestrated by the intracellular second messenger c-di-GMP and its receptor-effector FleQ. A bacterial enhancer binding protein, FleQ acts as a master regulator of both flagellar motility and adherence factor secretion and uses remarkably different transcription activation mechanisms depending on its dinucleotide loading state, adenosine triphosphatase (ATPase) activity, interactions with polymerase sigma (σ) factors, and complexation with a second ATPase, FleN. How the FleQ-FleN tandem can exert diverse effects through recognition of a conserved FleQ binding consensus has remained enigmatic. Here, we provide cryogenic electron microscopy (cryo-EM) structures of both c-di-GMP-bound and c-di-GMP-free FleQ-FleN complexes which deepen our understanding of the proteins' (di)nucleotide-dependent conformational switching and fine-tuned roles in gene expression regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17445.map.gz emd_17445.map.gz | 587.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17445-v30.xml emd-17445-v30.xml emd-17445.xml emd-17445.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17445_fsc.xml emd_17445_fsc.xml | 18.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17445.png emd_17445.png | 93.9 KB | ||
| Filedesc metadata |  emd-17445.cif.gz emd-17445.cif.gz | 6.8 KB | ||
| Others |  emd_17445_additional_1.map.gz emd_17445_additional_1.map.gz emd_17445_half_map_1.map.gz emd_17445_half_map_1.map.gz emd_17445_half_map_2.map.gz emd_17445_half_map_2.map.gz | 328.7 MB 621 MB 621 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17445 http://ftp.pdbj.org/pub/emdb/structures/EMD-17445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17445 | HTTPS FTP |
-Related structure data
| Related structure data |  8p53MC  8pb9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17445.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17445.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened cryo-EM density map of the c-di-GMP-free FleQ-FleN master regulator complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
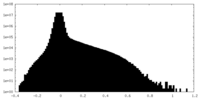
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened cryo-EM density map of the c-di-GMP-free FleQ-FleN...
| File | emd_17445_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened cryo-EM density map of the c-di-GMP-free FleQ-FleN master regulator complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
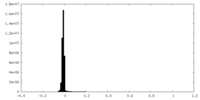
| Density Histograms |
-Half map: Half-map of the c-di-GMP-free FleQ-FleN master regulator complex
| File | emd_17445_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of the c-di-GMP-free FleQ-FleN master regulator complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
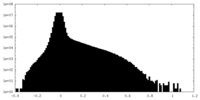
| Density Histograms |
-Half map: Half-map of the c-di-GMP-free FleQ-FleN master regulator complex
| File | emd_17445_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of the c-di-GMP-free FleQ-FleN master regulator complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : C-di-GMP-free FleQ-FleN complex
| Entire | Name: C-di-GMP-free FleQ-FleN complex |
|---|---|
| Components |
|
-Supramolecule #1: C-di-GMP-free FleQ-FleN complex
| Supramolecule | Name: C-di-GMP-free FleQ-FleN complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Full-length FleQ co-purified with His-tagged FleN D48A |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 288 KDa |
-Macromolecule #1: Antiactivator FleN
| Macromolecule | Name: Antiactivator FleN / type: protein_or_peptide / ID: 1 / Details: P. aeruginosa FleN D48A / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 |
| Molecular weight | Theoretical: 33.236332 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYYHHHHHH DYDIPTTLEV LFQGPMGSKQ MGSMHPVQVI AVTGGKGGVG KTNVSVNLAL ALADLGRRVM LLDAALGLAN VDVLLGLTP KRTLADVIEG RCELRDVLLL GPGGVRIVPA ASGTQSMVHL SPMQHAGLIQ AFSDISDNLD VLVVDTAAGI G DSVVSFVR ...String: MSYYHHHHHH DYDIPTTLEV LFQGPMGSKQ MGSMHPVQVI AVTGGKGGVG KTNVSVNLAL ALADLGRRVM LLDAALGLAN VDVLLGLTP KRTLADVIEG RCELRDVLLL GPGGVRIVPA ASGTQSMVHL SPMQHAGLIQ AFSDISDNLD VLVVDTAAGI G DSVVSFVR AAQEVLLVVC DEPTSITDAY ALIKLLNRDH GMTRFRVLAN MAHSPQEGRN LFAKLTKVTD RFLDVALQYV GV IPYDESV RKAVQKQRAV YEAFPRSKAS LAFKAVAQKV DSWPLPANPR GHLEFFVERL VQHPATGSAV UniProtKB: Antiactivator FleN |
-Macromolecule #2: Transcriptional regulator FleQ
| Macromolecule | Name: Transcriptional regulator FleQ / type: protein_or_peptide / ID: 2 Details: ...Details: MGSWRETKLLLIDDNLDRSRDLAVILNFLGEDQLTCNSEDWREVAAGLSNSREALCVLLGSVESKGGAVELLKQLASWDEYLPILLIGEPAPADWPEELRRRVLASLEMPPSYNKLLDSLHRAQVYREMYDQARERGRSREPNLFRSLVGTSRAIQQVRQMMQQVADTDASVLILGESGTGKEVVARNLHYHSKRREGPFVPVNCGAIPAELLESELFGHEKGAFTGAITSRAGRFELANGGTLFLDEIGDMPLPMQVKLLRVLQERTFERVGSNKTQNVDVRIIAATHKNLEKMIEDGTFREDLYYRLNVFPIEMAPLRERVEDIALLLNELISRMEHEKRGSIRFNSAAIMSLCRHDWPGNVRELANLVERLAIMHPYGVIGVGELPKKFRHVDDEDEQLASSLREELEERAAINAGLPGMDAPAMLPAEGLDLKDYLANLEQGLIQQALDDAGGVVARAAERLRIRRTTLVEKMRKYGMSRRDDDLSDD Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 55.494125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSWRETKLL LIDDNLDRSR DLAVILNFLG EDQLTCNSED WREVAAGLSN SREALCVLLG SVESKGGAVE LLKQLASWDE YLPILLIGE PAPADWPEEL RRRVLASLEM PPSYNKLLDS LHRAQVYREM YDQARERGRS REPNLFRSLV GTSRAIQQVR Q MMQQVADT ...String: MGSWRETKLL LIDDNLDRSR DLAVILNFLG EDQLTCNSED WREVAAGLSN SREALCVLLG SVESKGGAVE LLKQLASWDE YLPILLIGE PAPADWPEEL RRRVLASLEM PPSYNKLLDS LHRAQVYREM YDQARERGRS REPNLFRSLV GTSRAIQQVR Q MMQQVADT DASVLILGES GTGKEVVARN LHYHSKRREG PFVPVNCGAI PAELLESELF GHEKGAFTGA ITSRAGRFEL AN GGTLFLD EIGDMPLPMQ VKLLRVLQER TFERVGSNKT QNVDVRIIAA THKNLEKMIE DGTFREDLYY RLNVFPIEMA PLR ERVEDI ALLLNELISR MEHEKRGSIR FNSAAIMSLC RHDWPGNVRE LANLVERLAI MHPYGVIGVG ELPKKFRHVD DEDE QLASS LREELEERAA INAGLPGMDA PAMLPAEGLD LKDYLANLEQ GLIQQALDDA GGVVARAAER LRIRRTTLVE KMRKY GMSR RDDDLSDD UniProtKB: Transcriptional regulator FleQ |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: ACP |
|---|---|
| Molecular weight | Theoretical: 505.208 Da |
| Chemical component information |  ChemComp-ACP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM HEPES pH 8.0, 250mM NaCl, 2mM MgCl2, and 2% glycerol |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 21233 / Average electron dose: 51.5 e/Å2 / Details: 17099 micrographs retained for processing |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.3 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8p53: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)