+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Apo Malacoceros FaNaC1 in lipid nanodiscs | ||||||||||||
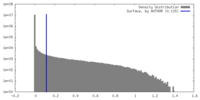
 Map data Map data | deepEMhancer postprocessed map of Malacoceros FaNaC1 in apo conformation used for model building and figure preparation | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Neuropeptide / ion channel / DEG/ENaC / MEMBRANE PROTEIN | ||||||||||||
| Biological species |  Malacoceros fuliginosus (invertebrata) Malacoceros fuliginosus (invertebrata) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||
 Authors Authors | Kalienkova V / Dandamudi M / Paulino C / Lynagh T | ||||||||||||
| Funding support |  Netherlands, Netherlands,  Norway, Norway,  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis for excitatory neuropeptide signaling. Authors: Valeria Kalienkova / Mowgli Dandamudi / Cristina Paulino / Timothy Lynagh /    Abstract: Rapid signaling between neurons is mediated by ligand-gated ion channels, cell-surface proteins with an extracellular ligand-binding domain and a membrane-spanning ion channel domain. The ...Rapid signaling between neurons is mediated by ligand-gated ion channels, cell-surface proteins with an extracellular ligand-binding domain and a membrane-spanning ion channel domain. The degenerin/epithelial sodium channel (DEG/ENaC) superfamily is diverse in terms of its gating stimuli, with some DEG/ENaCs gated by neuropeptides, and others gated by pH, mechanical force or enzymatic activity. The mechanism by which ligands bind to and activate DEG/ENaCs is poorly understood. Here we dissected the structural basis for neuropeptide-gated activity of a neuropeptide-gated DEG/ENaC, FMRFamide-gated sodium channel 1 (FaNaC1) from the annelid worm Malacoceros fuliginosus, using cryo-electron microscopy. Structures of FaNaC1 in the ligand-free resting state and in several ligand-bound states reveal the ligand-binding site and capture the ligand-induced conformational changes of channel gating, which we verified with complementary mutagenesis experiments. Our results illuminate channel gating in DEG/ENaCs and offer a structural template for experimental dissection of channel pharmacology and ion conduction. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16982.map.gz emd_16982.map.gz | 36.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16982-v30.xml emd-16982-v30.xml emd-16982.xml emd-16982.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16982.png emd_16982.png | 101.9 KB | ||
| Masks |  emd_16982_msk_1.map emd_16982_msk_1.map | 40.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16982.cif.gz emd-16982.cif.gz | 7 KB | ||
| Others |  emd_16982_additional_1.map.gz emd_16982_additional_1.map.gz emd_16982_half_map_1.map.gz emd_16982_half_map_1.map.gz emd_16982_half_map_2.map.gz emd_16982_half_map_2.map.gz | 30.8 MB 30.9 MB 31 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16982 http://ftp.pdbj.org/pub/emdb/structures/EMD-16982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16982 | HTTPS FTP |
-Validation report
| Summary document |  emd_16982_validation.pdf.gz emd_16982_validation.pdf.gz | 645 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16982_full_validation.pdf.gz emd_16982_full_validation.pdf.gz | 644.6 KB | Display | |
| Data in XML |  emd_16982_validation.xml.gz emd_16982_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_16982_validation.cif.gz emd_16982_validation.cif.gz | 13.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16982 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16982 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16982 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16982 | HTTPS FTP |
-Related structure data
| Related structure data |  8on8MC  8on7C  8on9C  8onaC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16982.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16982.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer postprocessed map of Malacoceros FaNaC1 in apo conformation used for model building and figure preparation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.022 Å | ||||||||||||||||||||||||||||||||||||
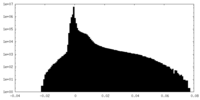
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16982_msk_1.map emd_16982_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
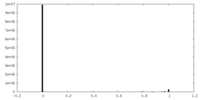
| Density Histograms |
-Additional map: refined map of Malacoceros FaNaC1 in apo conformation...
| File | emd_16982_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refined map of Malacoceros FaNaC1 in apo conformation used in model refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
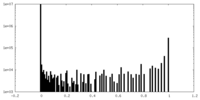
| Density Histograms |
-Half map: halfmap1 of Malacoceros FaNaC1 in apo conformation
| File | emd_16982_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 of Malacoceros FaNaC1 in apo conformation | ||||||||||||
| Projections & Slices |
| ||||||||||||
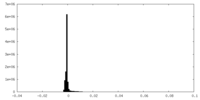
| Density Histograms |
-Half map: halfmap2 of Malacoceros FaNaC1 in apo conformation
| File | emd_16982_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 of Malacoceros FaNaC1 in apo conformation | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : FMRFamide-gated sodium channel 1 (FaNaC1)
| Entire | Name: FMRFamide-gated sodium channel 1 (FaNaC1) |
|---|---|
| Components |
|
-Supramolecule #1: FMRFamide-gated sodium channel 1 (FaNaC1)
| Supramolecule | Name: FMRFamide-gated sodium channel 1 (FaNaC1) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Malacoceros fuliginosus (invertebrata) Malacoceros fuliginosus (invertebrata) |
| Molecular weight | Theoretical: 205 KDa |
-Macromolecule #1: FMRFamide-gated sodium channel 1
| Macromolecule | Name: FMRFamide-gated sodium channel 1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Malacoceros fuliginosus (invertebrata) Malacoceros fuliginosus (invertebrata) |
| Molecular weight | Theoretical: 68.605836 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSAIRDVMTK FAEQTTMHGV PKVINAKSSM GRLFWSLVCL AAGAMFCLQM SEVLQRYFSY PKKVTVEVVP TPVPFPSISI CNMRNLDVH ILNTLNRMFI EDDRPFSNIN KSEHEFIRAY MKKVAKYAPL FWNYQDEYPE VFQEIFSRTT FSANIDPEVI A LAAVQLEG ...String: MSAIRDVMTK FAEQTTMHGV PKVINAKSSM GRLFWSLVCL AAGAMFCLQM SEVLQRYFSY PKKVTVEVVP TPVPFPSISI CNMRNLDVH ILNTLNRMFI EDDRPFSNIN KSEHEFIRAY MKKVAKYAPL FWNYQDEYPE VFQEIFSRTT FSANIDPEVI A LAAVQLEG FVVNCHYAGH RCNKTRDFYR FFDPYYFNCF TYKAHEPTDI EDNLSEGIEN GWSSILLSGS GMLDKNDEIR ML PGLHEWR SAVSASEGVR VVIHPPSTTP YPFTEGYDVP PGFSASFGIH PRRNIRIGPP HGNCSDKNPF GDGTERYRLM ACQ KMCMQH YIVETCGCAD VGLPKLPLQA NISWCRDDDN FPDECMFTAS EECLQLLMQL HNRIKCARSI KSKITKNTTA MEAC NCFPP CDEVSYDVSY SLSKWPSAGY EGDAAYFDVF GIEKFNERFN KTGTQGKYEL FTKYFNVSNR EESMKDFARL NVYIA DSNV VKTQESEDYT RNQLVSDIGG QLGLWVGISL ITLAEVLELI IDLFRLFSKH TYRSVPVIRQ SIKYKDKRNG AEMNYD TRY SQSNGGPHAR YLHHGHSIPK HPPELPDTSL ALEVLFQ |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.44 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: at 5 mA | |||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 288.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | nanodisc-reconstituted Malacoceros FaNaC1 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 90.0 K / Max: 105.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 2 / Number real images: 5361 / Average exposure time: 9.0 sec. / Average electron dose: 48.81 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.3 µm / Calibrated magnification: 49407 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



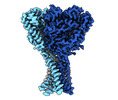



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)