+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Type six secretion system exported effector 5 (Tse5) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pore-forming protein / P.aeruginosa / effector / Bacterial Rearrangement hot spot protein / ion channel / type VI secretion system / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein secretion by the type VI secretion system / toxin sequestering activity Similarity search - Function | |||||||||
| Biological species |  Pseudomonas (RNA similarity group I) / Pseudomonas (RNA similarity group I) /  | |||||||||
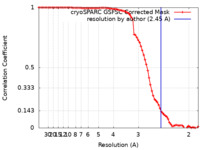
| Method | single particle reconstruction / cryo EM / Resolution: 2.45 Å | |||||||||
 Authors Authors | Gonzalez-Magana A / Tascon I / Ubarretxena-Belandia I / Albesa-Jove D | |||||||||
| Funding support |  Spain, 2 items Spain, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural and functional insights into the delivery of a bacterial Rhs pore-forming toxin to the membrane. Authors: Amaia González-Magaña / Igor Tascón / Jon Altuna-Alvarez / María Queralt-Martín / Jake Colautti / Carmen Velázquez / Maialen Zabala / Jessica Rojas-Palomino / Marité Cárdenas / ...Authors: Amaia González-Magaña / Igor Tascón / Jon Altuna-Alvarez / María Queralt-Martín / Jake Colautti / Carmen Velázquez / Maialen Zabala / Jessica Rojas-Palomino / Marité Cárdenas / Antonio Alcaraz / John C Whitney / Iban Ubarretxena-Belandia / David Albesa-Jové /   Abstract: Bacterial competition is a significant driver of toxin polymorphism, which allows continual compensatory evolution between toxins and the resistance developed to overcome their activity. Bacterial ...Bacterial competition is a significant driver of toxin polymorphism, which allows continual compensatory evolution between toxins and the resistance developed to overcome their activity. Bacterial Rearrangement hot spot (Rhs) proteins represent a widespread example of toxin polymorphism. Here, we present the 2.45 Å cryo-electron microscopy structure of Tse5, an Rhs protein central to Pseudomonas aeruginosa type VI secretion system-mediated bacterial competition. This structural insight, coupled with an extensive array of biophysical and genetic investigations, unravels the multifaceted functional mechanisms of Tse5. The data suggest that interfacial Tse5-membrane binding delivers its encapsulated pore-forming toxin fragment to the target bacterial membrane, where it assembles pores that cause cell depolarisation and, ultimately, bacterial death. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16778.map.gz emd_16778.map.gz | 26.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16778-v30.xml emd-16778-v30.xml emd-16778.xml emd-16778.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16778_fsc.xml emd_16778_fsc.xml | 7.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16778.png emd_16778.png | 84.7 KB | ||
| Masks |  emd_16778_msk_1.map emd_16778_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16778.cif.gz emd-16778.cif.gz | 7.1 KB | ||
| Others |  emd_16778_half_map_1.map.gz emd_16778_half_map_1.map.gz emd_16778_half_map_2.map.gz emd_16778_half_map_2.map.gz | 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16778 http://ftp.pdbj.org/pub/emdb/structures/EMD-16778 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16778 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16778 | HTTPS FTP |
-Validation report
| Summary document |  emd_16778_validation.pdf.gz emd_16778_validation.pdf.gz | 949 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16778_full_validation.pdf.gz emd_16778_full_validation.pdf.gz | 948.6 KB | Display | |
| Data in XML |  emd_16778_validation.xml.gz emd_16778_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_16778_validation.cif.gz emd_16778_validation.cif.gz | 20.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16778 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16778 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16778 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16778 | HTTPS FTP |
-Related structure data
| Related structure data |  8cp6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16778.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16778.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.921 Å | ||||||||||||||||||||||||||||||||||||
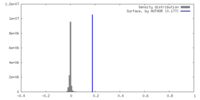
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16778_msk_1.map emd_16778_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
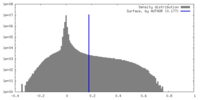
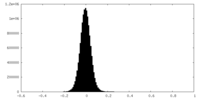
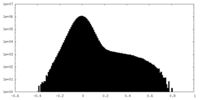
| Density Histograms |
-Half map: #2
| File | emd_16778_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
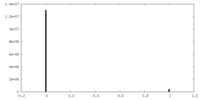
| Density Histograms |
-Half map: #1
| File | emd_16778_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
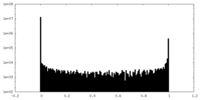
| Density Histograms |
- Sample components
Sample components
-Entire : Tse5
| Entire | Name: Tse5 |
|---|---|
| Components |
|
-Supramolecule #1: Tse5
| Supramolecule | Name: Tse5 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Pseudomonas aeruginosa type VI secretion system (T6SS) exported effector Tse5. The three chains result from the auto-cleavage of Tse5 |
|---|---|
| Source (natural) | Organism:  Pseudomonas (RNA similarity group I) Pseudomonas (RNA similarity group I) |
| Molecular weight | Theoretical: 146 KDa |
-Macromolecule #1: Toxin protein Tse5
| Macromolecule | Name: Toxin protein Tse5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.456322 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMGSSHHHHH HHHHSSGENL YFQGGSMSGL PVSHVGEKVS GGVISTGSPT VHVGSSAVGL ADRVSACVPL VGK UniProtKB: Toxin protein Tse5 |
-Macromolecule #2: Toxin protein Tse5
| Macromolecule | Name: Toxin protein Tse5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 125.970461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PVNPMLGSKL LPEEVDFALA APDTFTFARG YLSSNPRIGR LGRGWWLPGE SMHLELSEDA CVLVDAQGRR IGFPALAPGA QHYSGSEEL WLRRGGSSGG EAQAWRGRWA AVPAELQTQE GSVLVLSGHS YLHFQRCPDG IWRLQASFGR AGYRTEFRWS G RGLLTGVR ...String: PVNPMLGSKL LPEEVDFALA APDTFTFARG YLSSNPRIGR LGRGWWLPGE SMHLELSEDA CVLVDAQGRR IGFPALAPGA QHYSGSEEL WLRRGGSSGG EAQAWRGRWA AVPAELQTQE GSVLVLSGHS YLHFQRCPDG IWRLQASFGR AGYRTEFRWS G RGLLTGVR DSAGRSYALV YQQACEPSEG DDGLRLFGVI LASHDGPPPD YIDPQSPGLD WLVRYQFSDS GDLIAVRDRL GQ VVRVFAW REHMLVAHGE PGGLEVRYEW DVHAPHGRVV KQIEAGGLTR TFRYLRDATE VSDSLGRVER YEFAGEGGQR RWT ALVRAD GSRSEFDYDL FGRLVAMRDP LGRETRRRRD GQGRMLEEES PGKARYRKRV DEETGLLVEL EDAMQRRWTF ERDE RGNAT TVRGPAGSTR YAYEDPRLPD RPTRIVDPRG GERRLEWNRF GLLAALTDCS GQVWRYDYDN EGRLVASSDP LGQLT RRRY DPLGQLIGLE LADGSALSYE YDALGRQTRI ADAEGHATLF SWGHGDLLAR VSDAGGGELS YLHDEAGRLV ALTNEN GVQ AQFRYDLLDR LVEETGFDGR RQRYRYNAAD ELIAREDADG RETTYAYDRD GRLASIRVPA TEHAPALVER YRWLADG RL ASAGGADCEV RYTYDEVGNL RLESQVHADG WVYSVEHSHD ALGVRQTSRY GDAPPVAWLT YGPGHLHGAL VGAVELAF E RDALHREVRR DARRDGQDDA LFTQERQHAP LGRLQRSRLR LAGGFDWQRG YRYDGLGQLV GIDDNQYPSV RYEYDLGGR LLASRRAGAA ASTYRYDAAG NRLEGVGEHA REDARQAFAE NELYRSGFSR SETRASQAGE GPARWAGNRV ERIAGNRYRF DALGNLVER IGADGERLRL AYDGAQRLVH LTRDYADGTR LEARYRYDAL SRRIAKVVLR DGVEQQVRFG WDGDRQCAEA F ARELRTTV HEPGGFVPLL RLEQACEPDP PELLQLRQAF AAEGQPLPAQ CVPALGEARI AFFHTDHLGT PLQLSDERGQ LR WQGVPDD WRAVAPERQP GAQPIRFQGQ YHDEESGLYY NRYRYYLPEA GRYASQDPLG LGGGPNPYAY ALNAPTLAYD PTG L UniProtKB: Toxin protein Tse5 |
-Macromolecule #3: Toxin protein Tse5
| Macromolecule | Name: Toxin protein Tse5 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.592012 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: IIPLVVIGAF AARAAIGAAL GAGIELGMQT GKQVLGQMKD NWDSDRDLTD IKWKCIDINW KHVGASAAIG TVAPGMLSTG KTVVQSAKA IRTLSGQAAN TANRAAKLAA RKAAHADTIK KAVATQAAWQ TGKQIVKCPL KDEEEECPPQ UniProtKB: Toxin protein Tse5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 3.6 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 94 % / Chamber temperature: 289 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Number grids imaged: 1 / Number real images: 10244 / Average exposure time: 10.84 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: in silico model / Details: Buccanner |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8cp6: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)