+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Deinococcus radidurans HPI S-layer | |||||||||
 Map data Map data | PostProcessed map with B-factor sharpening. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | S-layer / cell wall organization / Prokaryotic membrane lipoprotein lipid attachment site profile. / extracellular region / Hexagonally packed intermediate-layer surface protein Function and homology information Function and homology information | |||||||||
| Biological species |  Deinococcus radiodurans (radioresistant) / Deinococcus radiodurans (radioresistant) /  Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422) (radioresistant) Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422) (radioresistant) | |||||||||
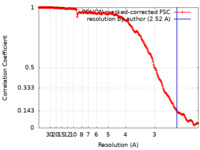
| Method | single particle reconstruction / cryo EM / Resolution: 2.52 Å | |||||||||
 Authors Authors | von Kuegelgen A / Yamashita K / Murshudov G / Bharat T | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Interdigitated immunoglobulin arrays form the hyperstable surface layer of the extremophilic bacterium . Authors: Andriko von Kügelgen / Sofie van Dorst / Keitaro Yamashita / Danielle L Sexton / Elitza I Tocheva / Garib Murshudov / Vikram Alva / Tanmay A M Bharat /    Abstract: is an atypical diderm bacterium with a remarkable ability to tolerate various environmental stresses, due in part to its complex cell envelope encapsulated within a hyperstable surface layer (S- ... is an atypical diderm bacterium with a remarkable ability to tolerate various environmental stresses, due in part to its complex cell envelope encapsulated within a hyperstable surface layer (S-layer). Despite decades of research on this cell envelope, atomic structural details of the S-layer have remained obscure. In this study, we report the electron cryomicroscopy structure of the S-layer, showing how it is formed by the Hexagonally Packed Intermediate-layer (HPI) protein arranged in a planar hexagonal lattice. The HPI protein forms an array of immunoglobulin-like folds within the S-layer, with each monomer extending into the adjacent hexamer, resulting in a highly interconnected, stable, sheet-like arrangement. Using electron cryotomography and subtomogram averaging from focused ion beam-milled cells, we have obtained a structure of the cellular S-layer, showing how this HPI S-layer coats native membranes on the surface of cells. Our S-layer structure from the diderm bacterium shows similarities to immunoglobulin-like domain-containing S-layers from monoderm bacteria and archaea, highlighting common features in cell surface organization across different domains of life, with connotations on the evolution of immunoglobulin-based molecular recognition systems in eukaryotes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16694.map.gz emd_16694.map.gz | 227.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16694-v30.xml emd-16694-v30.xml emd-16694.xml emd-16694.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16694_fsc.xml emd_16694_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16694.png emd_16694.png | 253.4 KB | ||
| Masks |  emd_16694_msk_1.map emd_16694_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_16694_half_map_1.map.gz emd_16694_half_map_1.map.gz emd_16694_half_map_2.map.gz emd_16694_half_map_2.map.gz | 217.3 MB 217.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16694 http://ftp.pdbj.org/pub/emdb/structures/EMD-16694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16694 | HTTPS FTP |
-Validation report
| Summary document |  emd_16694_validation.pdf.gz emd_16694_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16694_full_validation.pdf.gz emd_16694_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_16694_validation.xml.gz emd_16694_validation.xml.gz | 23.4 KB | Display | |
| Data in CIF |  emd_16694_validation.cif.gz emd_16694_validation.cif.gz | 31.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16694 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16694 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16694 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16694 | HTTPS FTP |
-Related structure data
| Related structure data |  8ckaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16694.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16694.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PostProcessed map with B-factor sharpening. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.092 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16694_msk_1.map emd_16694_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_16694_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_16694_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of Hexagonally Packed Intermediate-layer (HPI) protein
| Entire | Name: Structure of Hexagonally Packed Intermediate-layer (HPI) protein |
|---|---|
| Components |
|
-Supramolecule #1: Structure of Hexagonally Packed Intermediate-layer (HPI) protein
| Supramolecule | Name: Structure of Hexagonally Packed Intermediate-layer (HPI) protein type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Structure of Hexagonally Packed Intermediate-layer (HPI) protein |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans (radioresistant) / Location in cell: extracellular Deinococcus radiodurans (radioresistant) / Location in cell: extracellular |
-Macromolecule #1: Hexagonally packed intermediate-layer surface protein
| Macromolecule | Name: Hexagonally packed intermediate-layer surface protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422) (radioresistant) Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422) (radioresistant) |
| Molecular weight | Theoretical: 99.424 KDa |
| Sequence | String: MKKNIALMAL TGILTLASCG QNGTGTTPTA DACATANTCS VTVNISGVSS ADFDVTMDGK TTSMTLSNGQ KLPVAKTGTV TLTPKAKDG YTTPAAQSTT ISSTNLTPSV NFAYTTVPST GNGNGNGGTT PTQPFTLNIT SPTNGAAATT GTPIRVVFTS S VALSSATC ...String: MKKNIALMAL TGILTLASCG QNGTGTTPTA DACATANTCS VTVNISGVSS ADFDVTMDGK TTSMTLSNGQ KLPVAKTGTV TLTPKAKDG YTTPAAQSTT ISSTNLTPSV NFAYTTVPST GNGNGNGGTT PTQPFTLNIT SPTNGAAATT GTPIRVVFTS S VALSSATC KIGNSAAVNA QVSSTGGYCD VTPTTAGGGL ITVTGTANGQ TVSSTVTVDV KAPVVDNRYG TVTPAGDQEL TL TNEGIVK DADNGWRRLG QGVSTPSDPN GNVDIYVKGT VNFSVNAAAG SKVEVFLART TGSDVPTNDD VQAGDVLRSV AST SGTETF SLDSRRLAEF DGVRKWIVVR INGTQVTYQP VIADNKGPQQ PDPELNGVQN AYSNILNNYN NSGLTYVRGD VNVF TGNPS LQDREFGQAP LGSSFVQRRP SGFESIRYYL VPETAFGNKA LQESDEMLRA KAIKSVATVV SAPVLEPGTV KATSF SRVI GSGATSTVTP KAQDNVTYRV YAISRDQLGN ETASATYELV RFDNVGPTIT GSVIRDTSDL PFASQEPERC LSDIAT ITL GGITDNAGGV GLNPGQGLTF TLGGRQIQAG QFDTNQLADG EYTIGFNSLT DALGNPVVSA PTNAKVYIDN TDPTVNF NR AVMQGTFASG ERVSVESDAS DGGCGVYETR LFWDTDNGVV DDATTTPAIG HPVQFARQRV TDGAKADSLN AGWNALQL P NGAGAVYLRA LVVDRAGNAT ISTTPIVVNA KITNQARPLL GGFDAFKRNA SAQFMSNSNA ISGVNGTAVT PNTTANSAL DNILSLDSVG TLTTNAYLPR GATETAITEK IRNVGAYGRF DATQWNRIRD YQLNTDPTLR SAYVNAGNLA NQRGNNWRIR TPWVELGSS DTANTQQKFD FNSDLLNDFY FGRTFGNNDN VNLFSYDQFN GIVSGTAGAY SFYGETVQK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 50 mM HEPES/NaOH pH=7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM CaCl2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA | |||||||||||||||
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: absorption for 60 sec and blotted for 5 sec with blot force -10. | |||||||||||||||
| Details | In vitro isolate Hexagonally Packed Intermediate-layer (HPI) layer |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Spherical aberration corrector: not used / Chromatic aberration corrector: not used / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 1002 / Average exposure time: 3.43 sec. / Average electron dose: 53.245 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 2.0 µm / Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 19.48 / Target criteria: Best Fit |
|---|---|
| Output model |  PDB-8cka: |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X