[English] 日本語
 Yorodumi
Yorodumi- EMDB-16680: BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN | ||||||||||||
 Map data Map data | Sharpened map BA4/5 fab with BA4 Spike. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | SARS-CoV2 / spike / glycoprotein / coronavirus / antibody / fab / BA.4 / BA.5 / BA.4/5-5 / cross-protective / omicron variant / vaccine / therapeutic / complex / neutralising / convalescent sera / viral protein / immune system | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||
 Authors Authors | Duyvesteyn HME / Ren J / Stuart DI / Fry EE | ||||||||||||
| Funding support |  China, China,  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86. Authors: Daming Zhou / Piyada Supasa / Chang Liu / Aiste Dijokaite-Guraliuc / Helen M E Duyvesteyn / Muneeswaran Selvaraj / Alexander J Mentzer / Raksha Das / Wanwisa Dejnirattisai / Nigel Temperton ...Authors: Daming Zhou / Piyada Supasa / Chang Liu / Aiste Dijokaite-Guraliuc / Helen M E Duyvesteyn / Muneeswaran Selvaraj / Alexander J Mentzer / Raksha Das / Wanwisa Dejnirattisai / Nigel Temperton / Paul Klenerman / Susanna J Dunachie / Elizabeth E Fry / Juthathip Mongkolsapaya / Jingshan Ren / David I Stuart / Gavin R Screaton /    Abstract: Under pressure from neutralising antibodies induced by vaccination or infection the SARS-CoV-2 spike gene has become a hotspot for evolutionary change, leading to the failure of all mAbs developed ...Under pressure from neutralising antibodies induced by vaccination or infection the SARS-CoV-2 spike gene has become a hotspot for evolutionary change, leading to the failure of all mAbs developed for clinical use. Most potent antibodies bind to the receptor binding domain which has become heavily mutated. Here we study responses to a conserved epitope in sub-domain-1 (SD1) of spike which have become more prominent because of mutational escape from antibodies directed to the receptor binding domain. Some SD1 reactive mAbs show potent and broad neutralization of SARS-CoV-2 variants. We structurally map the dominant SD1 epitope and provide a mechanism of action by blocking interaction with ACE2. Mutations in SD1 have not been sustained to date, but one, E554K, leads to escape from mAbs. This mutation has now emerged in several sublineages including BA.2.86, reflecting selection pressure on the virus exerted by the increasing prominence of the anti-SD1 response. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16680.map.gz emd_16680.map.gz | 318.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16680-v30.xml emd-16680-v30.xml emd-16680.xml emd-16680.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16680.png emd_16680.png | 54.2 KB | ||
| Filedesc metadata |  emd-16680.cif.gz emd-16680.cif.gz | 7.5 KB | ||
| Others |  emd_16680_half_map_1.map.gz emd_16680_half_map_1.map.gz emd_16680_half_map_2.map.gz emd_16680_half_map_2.map.gz | 322.9 MB 322.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16680 http://ftp.pdbj.org/pub/emdb/structures/EMD-16680 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16680 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16680 | HTTPS FTP |
-Related structure data
| Related structure data |  8cinMC  8r1cC  8r1dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16680.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16680.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map BA4/5 fab with BA4 Spike. | ||||||||||||||||||||||||||||||||||||
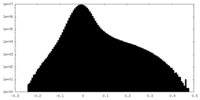
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7303 Å | ||||||||||||||||||||||||||||||||||||
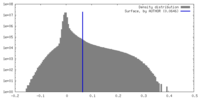
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map BA4/5 fab with BA4 Spike.
| File | emd_16680_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map BA4/5 fab with BA4 Spike. | ||||||||||||
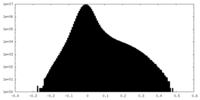
| Projections & Slices |
| ||||||||||||
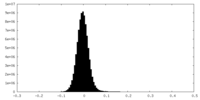
| Density Histograms |
-Half map: Half map BA4/5 fab with BA4 Spike.
| File | emd_16680_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map BA4/5 fab with BA4 Spike. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-Coronavirus-2 BA.4 Spike Glycoprotein in complex with BA.4/5...
| Entire | Name: SARS-Coronavirus-2 BA.4 Spike Glycoprotein in complex with BA.4/5-5 fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-Coronavirus-2 BA.4 Spike Glycoprotein in complex with BA.4/5...
| Supramolecule | Name: SARS-Coronavirus-2 BA.4 Spike Glycoprotein in complex with BA.4/5-5 fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: BA.4/5-5 fab heavy and light chains.
| Supramolecule | Name: BA.4/5-5 fab heavy and light chains. / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2-#3 Details: Fab fragment, recombinantely expressed (sequence from convalescent sera). |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: SARS-Coronavirus-2 BA.4 Spike Glycoprotein
| Supramolecule | Name: SARS-Coronavirus-2 BA.4 Spike Glycoprotein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 124.616305 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TQSYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRGW IFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLDV YYHKNNKSWM ESEFRVYSSA NNCTFEYVSQ PFLMDLEGKQ G NFKNLREF ...String: TQSYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRGW IFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLDV YYHKNNKSWM ESEFRVYSSA NNCTFEYVSQ PFLMDLEGKQ G NFKNLREF VFKNIDGYFK IYSKHTPINL VRDLPQGFSA LEPLVDLPIG INITRFQTLL ALHRSYLTPG DSSSGWTAGA AA YYVGYLQ PRTFLLKYNE NGTITDAVDC ALDPLSETKC TLKSFTVEKG IYQTSNFRVQ PTESIVRFPN ITNLCPFDEV FNA TRFASV YAWNRKRISN CVADYSVLYN FAPFFAFKCY GVSPTKLNDL CFTNVYADSF VIRGNEVSQI APGQTGNIAD YNYK LPDDF TGCVIAWNSN KLDSKVGGNY NYRYRLFRKS NLKPFERDIS TEIYQAGNKP CNGVAGVNCY FPLQSYGFRP TYGVG HQPY RVVVLSFELL HAPATVCGPK KSTNLVKNKC VNFNFNGLTG TGVLTESNKK FLPFQQFGRD IADTTDAVRD PQTLEI LDI TPCSFGGVSV ITPGTNTSNQ VAVLYQGVNC TEVPVAIHAD QLTPTWRVYS TGSNVFQTRA GCLIGAEYVN NSYECDI PI GAGICASYQT QTNSPRRARS VASQSIIAYT MSLGAENSVA YSNNSIAIPT NFTISVTTEI LPVSMTKTSV DCTMYICG D STECSNLLLQ YGSFCTQLKR ALTGIAVEQD KNTQEVFAQV KQIYKTPPIK YFGGFNFSQI LPDPSKPSKR SFIEDLLFN KVTLADAGFI KQYGDCLGDI AARDLICAQK FNGLTVLPPL LTDEMIAQYT SALLAGTITS GWTFGAGAAL QIPFAMQMAY RFNGIGVTQ NVLYENQKLI ANQFNSAIGK IQDSLSSTAS ALGKLQDVVN HNAQALNTLV KQLSSKFGAI SSVLNDILSR L DPPEAEVQ IDRLITGRLQ SLQTYVTQQL IRAAEIRASA NLAATKMSEC VLGQSKRVDF CGKGYHLMSF PQSAPHGVVF LH VTYVPAQ EKNFTTAPAI CHDGKAHFPR EGVFVSNGTH WFVTQRNFYE PQIITTDNTF VSGNCDVVIG IVNNTVYDPL QPE LDS UniProtKB: Spike glycoprotein |
-Macromolecule #2: BA.4/5-5 fab HEAVY CHAIN
| Macromolecule | Name: BA.4/5-5 fab HEAVY CHAIN / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.612451 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGPG LVKPSETLSL TCSVSGGSID NFYWSWIRQP PGKGLEWIGN IYYGGSGNYN PSLKSRVTIS VDTSKNQVSL KLRSVTAAD TAVYYCARDS VWYTGSYGLI YWGPGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT ...String: QVQLVESGPG LVKPSETLSL TCSVSGGSID NFYWSWIRQP PGKGLEWIGN IYYGGSGNYN PSLKSRVTIS VDTSKNQVSL KLRSVTAAD TAVYYCARDS VWYTGSYGLI YWGPGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKR VEPKS |
-Macromolecule #3: BA.4/5-5 fab LIGHT CHAIN
| Macromolecule | Name: BA.4/5-5 fab LIGHT CHAIN / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.616102 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SVVTQPASVS GSPGQSITIS CTGTSSDVGS YNLVSWFQQH PGKAPKLMIY EVTKRPSGIS NRFSGSKSGN TASLTISRLQ AEDEADYYC CSYAGSSTVV FGGGTKLTVL GQPKANPTVT LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADSSPVK A GVETTTPS ...String: SVVTQPASVS GSPGQSITIS CTGTSSDVGS YNLVSWFQQH PGKAPKLMIY EVTKRPSGIS NRFSGSKSGN TASLTISRLQ AEDEADYYC CSYAGSSTVV FGGGTKLTVL GQPKANPTVT LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADSSPVK A GVETTTPS KQSNNKYAAS SYLSLTPEQW KSHRSYSCQV THEGSTVEKT VAPTEC |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.5 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 65.6 |
|---|---|
| Output model |  PDB-8cin: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)