[English] 日本語
 Yorodumi
Yorodumi- EMDB-16564: Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium and 3'-phosphoadenosine-5'-phosphosulfate | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Deacetylase / Sulfotransferase / Heparan Sulfate / Carbohydrate / Glycosaminoglycan | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information[heparan sulfate]-glucosamine N-sulfotransferase / heparan sulfate N-sulfotransferase activity / heparan sulfate N-deacetylase activity / N-acetylglucosamine deacetylase activity / heparin proteoglycan biosynthetic process / embryonic neurocranium morphogenesis / embryonic viscerocranium morphogenesis / HS-GAG biosynthesis / cardiac septum development / glycosaminoglycan metabolic process ...[heparan sulfate]-glucosamine N-sulfotransferase / heparan sulfate N-sulfotransferase activity / heparan sulfate N-deacetylase activity / N-acetylglucosamine deacetylase activity / heparin proteoglycan biosynthetic process / embryonic neurocranium morphogenesis / embryonic viscerocranium morphogenesis / HS-GAG biosynthesis / cardiac septum development / glycosaminoglycan metabolic process / heparan sulfate proteoglycan biosynthetic process / deacetylase activity / respiratory gaseous exchange by respiratory system / polysaccharide biosynthetic process / coronary vasculature development / positive regulation of smoothened signaling pathway / Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides / aorta development / midbrain development / forebrain development / fibroblast growth factor receptor signaling pathway / trans-Golgi network membrane / cell population proliferation / positive regulation of MAPK cascade / inflammatory response / Golgi membrane / Golgi apparatus Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
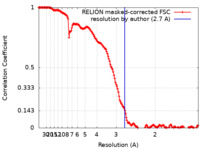
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||||||||
 Authors Authors | Mycroft-West CJ / Wu L | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural and mechanistic characterization of bifunctional heparan sulfate N-deacetylase-N-sulfotransferase 1. Authors: Courtney J Mycroft-West / Sahar Abdelkarim / Helen M E Duyvesteyn / Neha S Gandhi / Mark A Skidmore / Raymond J Owens / Liang Wu /    Abstract: Heparan sulfate (HS) polysaccharides are major constituents of the extracellular matrix, which are involved in myriad structural and signaling processes. Mature HS polysaccharides contain complex, ...Heparan sulfate (HS) polysaccharides are major constituents of the extracellular matrix, which are involved in myriad structural and signaling processes. Mature HS polysaccharides contain complex, non-templated patterns of sulfation and epimerization, which mediate interactions with diverse protein partners. Complex HS modifications form around initial clusters of glucosamine-N-sulfate (GlcNS) on nascent polysaccharide chains, but the mechanistic basis underpinning incorporation of GlcNS itself into HS remains unclear. Here, we determine cryo-electron microscopy structures of human N-deacetylase-N-sulfotransferase (NDST)1, the bifunctional enzyme primarily responsible for initial GlcNS modification of HS. Our structures reveal the architecture of both NDST1 deacetylase and sulfotransferase catalytic domains, alongside a non-catalytic N-terminal domain. The two catalytic domains of NDST1 adopt a distinct back-to-back topology that limits direct cooperativity. Binding analyses, aided by activity-modulating nanobodies, suggest that anchoring of the substrate at the sulfotransferase domain initiates the NDST1 catalytic cycle, providing a plausible mechanism for cooperativity despite spatial domain separation. Our data shed light on key determinants of NDST1 activity, and describe tools to probe NDST1 function in vitro and in vivo. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16564.map.gz emd_16564.map.gz | 301.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16564-v30.xml emd-16564-v30.xml emd-16564.xml emd-16564.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16564_fsc.xml emd_16564_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16564.png emd_16564.png | 61.8 KB | ||
| Masks |  emd_16564_msk_1.map emd_16564_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16564.cif.gz emd-16564.cif.gz | 6.3 KB | ||
| Others |  emd_16564_half_map_1.map.gz emd_16564_half_map_1.map.gz emd_16564_half_map_2.map.gz emd_16564_half_map_2.map.gz | 301.4 MB 301.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16564 http://ftp.pdbj.org/pub/emdb/structures/EMD-16564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16564 | HTTPS FTP |
-Related structure data
| Related structure data |  8ccyMC  8cd0C  8chsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16564.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16564.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
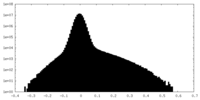
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16564_msk_1.map emd_16564_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
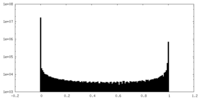
| Density Histograms |
-Half map: #2
| File | emd_16564_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
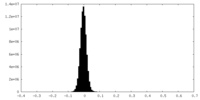
| Density Histograms |
-Half map: #1
| File | emd_16564_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
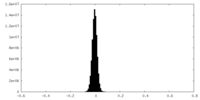
| Density Histograms |
- Sample components
Sample components
-Entire : Human heparan sulfate N-deacetylase-N-sulfotransferase 1
| Entire | Name: Human heparan sulfate N-deacetylase-N-sulfotransferase 1 |
|---|---|
| Components |
|
-Supramolecule #1: Human heparan sulfate N-deacetylase-N-sulfotransferase 1
| Supramolecule | Name: Human heparan sulfate N-deacetylase-N-sulfotransferase 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1
| Macromolecule | Name: Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.407461 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: SRTDPLVLVF VESLYSQLGQ EVVAILESSR FKYRTEIAPG KGDMPTLTDK GRGRFALIIY ENILKYVNLD AWNRELLDKY CVAYGVGII GFFKANENSL LSAQLKGFPL FLHSNLGLKD CSINPKSPLL YVTRPSEVEK GVLPGEDWTV FQSNHSTYEP V LLAKTRSS ...String: SRTDPLVLVF VESLYSQLGQ EVVAILESSR FKYRTEIAPG KGDMPTLTDK GRGRFALIIY ENILKYVNLD AWNRELLDKY CVAYGVGII GFFKANENSL LSAQLKGFPL FLHSNLGLKD CSINPKSPLL YVTRPSEVEK GVLPGEDWTV FQSNHSTYEP V LLAKTRSS ESIPHLGADA GLHAALHATV VQDLGLHDGI QRVLFGNNLN FWLHKLVFVD AVAFLTGKRL SLPLDRYILV DI DDIFVGK EGTRMKVEDV KALFDTQNEL RAHIPNFTFN LGYSGKFFHT GTNAEDAGDD LLLSYVKEFW WFPHMWSHMQ PHL FHNQSV LAEQMALNKK FAVEHGIPTD MGYAVAPHHS GVYPVHVQLY EAWKQVWSIR VTSTEEYPHL KPARYRRGFI HNGI MVLPR QTCGLFTHTI FYNEYPGGSS ELDKIINGGE LFLTVLLNPI SIFMTHLSNY GNDRLGLYTF KHLVRFLHSW TNLRL QTLP PVQLAQKYFQ IFSEEKDPLW QDPCEDKRHK DIWSKEKTCD RFPKLLIIGP QKTGTTALYL FLGMHPDLSS NYPSSE TFE EIQFFNGHNY HKGIDWYMEF FPIPSNTTSD FYFEKSANYF DSEVAPRRAA ALLPKAKVLT ILINPADRAY SWYQHQR AH DDPVALKYTF HEVITAGSDA SSKLRALQNR CLVPGWYATH IERWLSAYHA NQILVLDGKL LRTEPAKVMD MVQKFLGV T NTIDYHKTLA FDPKKGFWCQ LLEGGKTKCL GKSKGRKYPE MDLDSRAFLK DYYRDHNIEL SKLLYKMGQT LPTWLREDL QNTR UniProtKB: Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 |
-Macromolecule #2: ADENOSINE-3'-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-3'-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: A3P |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-A3P: |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 23 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Vitrification | Cryogen name: ETHANE |
| Details | Monodisperse sample from size exclusion |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 165000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)