[English] 日本語
 Yorodumi
Yorodumi- EMDB-16512: MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | ADDomer / VLP / Viral / like / Particle / AD3 / Penton / base / vaccine / COVID-19 / COVID / MINI-COVID / Dodecahedron / pb / protein / adenovirus / human / VIRUS LIKE PARTICLE | |||||||||||||||||||||||||||
| Function / homology | Adenovirus penton base protein / Adenovirus penton base protein / T=25 icosahedral viral capsid / endocytosis involved in viral entry into host cell / virion attachment to host cell / host cell nucleus / structural molecule activity / metal ion binding / Penton protein Function and homology information Function and homology information | |||||||||||||||||||||||||||
| Biological species |  Human adenovirus B3 Human adenovirus B3 | |||||||||||||||||||||||||||
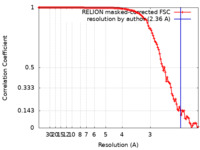
| Method | single particle reconstruction / cryo EM / Resolution: 2.36 Å | |||||||||||||||||||||||||||
 Authors Authors | Bufton JC / Capin J / Boruku U / Garzoni F / Schaffitzel C / Berger I | |||||||||||||||||||||||||||
| Funding support |  United Kingdom, 8 items United Kingdom, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Antib Ther / Year: 2023 Journal: Antib Ther / Year: 2023Title: generated antibodies guide thermostable ADDomer nanoparticle design for nasal vaccination and passive immunization against SARS-CoV-2. Authors: Dora Buzas / Adrian H Bunzel / Oskar Staufer / Emily J Milodowski / Grace L Edmunds / Joshua C Bufton / Beatriz V Vidana Mateo / Sathish K N Yadav / Kapil Gupta / Charlotte Fletcher / Maia K ...Authors: Dora Buzas / Adrian H Bunzel / Oskar Staufer / Emily J Milodowski / Grace L Edmunds / Joshua C Bufton / Beatriz V Vidana Mateo / Sathish K N Yadav / Kapil Gupta / Charlotte Fletcher / Maia K Williamson / Alexandra Harrison / Ufuk Borucu / Julien Capin / Ore Francis / Georgia Balchin / Sophie Hall / Mirella V Vega / Fabien Durbesson / Srikanth Lingappa / Renaud Vincentelli / Joe Roe / Linda Wooldridge / Rachel Burt / Ross J L Anderson / Adrian J Mulholland / Bristol Uncover Group / Jonathan Hare / Mick Bailey / Andrew D Davidson / Adam Finn / David Morgan / Jamie Mann / Joachim Spatz / Frederic Garzoni / Christiane Schaffitzel / Imre Berger /    Abstract: BACKGROUND: Due to COVID-19, pandemic preparedness emerges as a key imperative, necessitating new approaches to accelerate development of reagents against infectious pathogens. METHODS: Here, we developed an integrated approach combining synthetic, computational and structural methods with antibody selection and immunization to design, produce and validate nature-inspired ...METHODS: Here, we developed an integrated approach combining synthetic, computational and structural methods with antibody selection and immunization to design, produce and validate nature-inspired nanoparticle-based reagents against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). RESULTS: Our approach resulted in two innovations: (i) a thermostable nasal vaccine called ADDoCoV, displaying multiple copies of a SARS-CoV-2 receptor binding motif derived epitope and (ii) a ...RESULTS: Our approach resulted in two innovations: (i) a thermostable nasal vaccine called ADDoCoV, displaying multiple copies of a SARS-CoV-2 receptor binding motif derived epitope and (ii) a multivalent nanoparticle superbinder, called Gigabody, against SARS-CoV-2 including immune-evasive variants of concern (VOCs). generated neutralizing nanobodies and electron cryo-microscopy established authenticity and accessibility of epitopes displayed by ADDoCoV. Gigabody comprising multimerized nanobodies prevented SARS-CoV-2 virion attachment with picomolar EC. Vaccinating mice resulted in antibodies cross-reacting with VOCs including Delta and Omicron. CONCLUSION: Our study elucidates Adenovirus-derived dodecamer (ADDomer)-based nanoparticles for use in active and passive immunization and provides a blueprint for crafting reagents to combat ...CONCLUSION: Our study elucidates Adenovirus-derived dodecamer (ADDomer)-based nanoparticles for use in active and passive immunization and provides a blueprint for crafting reagents to combat respiratory viral infections. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16512.map.gz emd_16512.map.gz | 116.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16512-v30.xml emd-16512-v30.xml emd-16512.xml emd-16512.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16512_fsc.xml emd_16512_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_16512.png emd_16512.png | 60.5 KB | ||
| Filedesc metadata |  emd-16512.cif.gz emd-16512.cif.gz | 6.3 KB | ||
| Others |  emd_16512_half_map_1.map.gz emd_16512_half_map_1.map.gz emd_16512_half_map_2.map.gz emd_16512_half_map_2.map.gz | 117.6 MB 117.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16512 http://ftp.pdbj.org/pub/emdb/structures/EMD-16512 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16512 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16512 | HTTPS FTP |
-Related structure data
| Related structure data |  8c9nMC  11730 M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16512.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16512.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
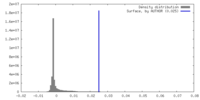
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16512_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
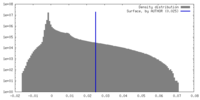
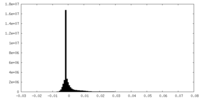
| Density Histograms |
-Half map: #2
| File | emd_16512_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
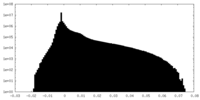
| Density Histograms |
- Sample components
Sample components
-Entire : ADDomer Mini-COVID viral like particle vaccine derived from Human...
| Entire | Name: ADDomer Mini-COVID viral like particle vaccine derived from Human Ad3 penton base protein. |
|---|---|
| Components |
|
-Supramolecule #1: ADDomer Mini-COVID viral like particle vaccine derived from Human...
| Supramolecule | Name: ADDomer Mini-COVID viral like particle vaccine derived from Human Ad3 penton base protein. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Human adenovirus B3 Human adenovirus B3 |
| Molecular weight | Theoretical: 3.9930516 MDa |
-Macromolecule #1: Penton protein
| Macromolecule | Name: Penton protein / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human adenovirus B3 Human adenovirus B3 |
| Molecular weight | Theoretical: 66.618547 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MRRRAVLGGA VVYPEGPPPS YESVMQQQAA MIQPPLEAPF VPPRYLAPTE GRNSIRYSEL SPLYDTTKLY LVDNKSADIA SLNYQNDHS NFLTTVVQNN DFTPTEASTQ TINFDERSRW GGQLKTIMHT NMPNVNEYMF SNKFKARVMV SRKAPEGEFY Q AGSTPCNG ...String: MRRRAVLGGA VVYPEGPPPS YESVMQQQAA MIQPPLEAPF VPPRYLAPTE GRNSIRYSEL SPLYDTTKLY LVDNKSADIA SLNYQNDHS NFLTTVVQNN DFTPTEASTQ TINFDERSRW GGQLKTIMHT NMPNVNEYMF SNKFKARVMV SRKAPEGEFY Q AGSTPCNG VEGFNCYFPL QSYGFQPTNG VGYGPVNDTY DHKEDILKYE WFEFILPEGN FSATMTIDLM NNAIIDNYLE IG RQNGVLE SDIGVKFDTR NFRLGWDPET KLIMPGVYTY EAFHPDIVLL PGCGVDFTES RLSNLLGIRK RHPFQEGFKI MYE DLEGGN IPALLDVTAY EESKKDTTTA RETTTLAVAE ETSEDVDDDI TRGDTYITEL EKQKREAAAA EVSRKKELKI QPLE KDSKS RSYNVLEDKI NTAYRSWYLS YNYGNPEKGI RSWTLLTTSD VTCGAEQVYW SLPDMMQDPV TFRSTRQVNN YPVVG AELM PVFSKSFYNE QAVYSQQLRQ ATSLTHVFNR FPENQILIRP PAPTITTVSE NVPALTDHGT LPLRSSIRGV QRVTVT DAR RRTCPYVYKA LGIVAPRVLS SRTF UniProtKB: Penton protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.1 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)