+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AQP7_inhibitor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | aquaglyceroporin / glycerol channel / dimer of tetramers / inhibitor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTransport of glycerol from adipocytes to the liver by Aquaporins / Passive transport by Aquaporins / glycerol channel activity / urea transmembrane transporter activity / glycerol transmembrane transport / water transport / water channel activity / lipid droplet / cytoplasmic vesicle membrane / cell-cell junction ...Transport of glycerol from adipocytes to the liver by Aquaporins / Passive transport by Aquaporins / glycerol channel activity / urea transmembrane transporter activity / glycerol transmembrane transport / water transport / water channel activity / lipid droplet / cytoplasmic vesicle membrane / cell-cell junction / basolateral plasma membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Huang P / Venskutonyte R / Gourdon P / Lindkvist-Petersson K | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Molecular basis for human aquaporin inhibition. Authors: Peng Huang / Hannah Åbacka / Carter J Wilson / Malene Lykke Wind / Michael Rűtzler / Anna Hagström-Andersson / Pontus Gourdon / Bert L de Groot / Raminta Venskutonytė / Karin Lindkvist-Petersson /    Abstract: Cancer invasion and metastasis are known to be potentiated by the expression of aquaporins (AQPs). Likewise, the expression levels of AQPs have been shown to be prognostic for survival in patients ...Cancer invasion and metastasis are known to be potentiated by the expression of aquaporins (AQPs). Likewise, the expression levels of AQPs have been shown to be prognostic for survival in patients and have a role in tumor growth, edema, angiogenesis, and tumor cell migration. Thus, AQPs are key players in cancer biology and potential targets for drug development. Here, we present the single-particle cryo-EM structure of human AQP7 at 3.2-Å resolution in complex with the specific inhibitor compound Z433927330. The structure in combination with MD simulations shows that the inhibitor binds to the endofacial side of AQP7. In addition, cancer cells treated with Z433927330 show reduced proliferation. The data presented here serve as a framework for the development of AQP inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16510.map.gz emd_16510.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16510-v30.xml emd-16510-v30.xml emd-16510.xml emd-16510.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16510_fsc.xml emd_16510_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16510.png emd_16510.png | 31.5 KB | ||
| Masks |  emd_16510_msk_1.map emd_16510_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16510.cif.gz emd-16510.cif.gz | 6.3 KB | ||
| Others |  emd_16510_half_map_1.map.gz emd_16510_half_map_1.map.gz emd_16510_half_map_2.map.gz emd_16510_half_map_2.map.gz | 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16510 http://ftp.pdbj.org/pub/emdb/structures/EMD-16510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16510 | HTTPS FTP |
-Related structure data
| Related structure data |  8c9hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16510.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16510.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8464 Å | ||||||||||||||||||||||||||||||||||||
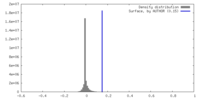
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16510_msk_1.map emd_16510_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
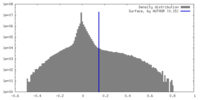
| Density Histograms |
-Half map: #2
| File | emd_16510_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
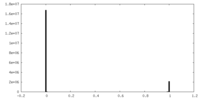
| Density Histograms |
-Half map: #1
| File | emd_16510_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
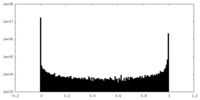
| Density Histograms |
- Sample components
Sample components
-Entire : AQP7 dimer of tetramers with inhibitor
| Entire | Name: AQP7 dimer of tetramers with inhibitor |
|---|---|
| Components |
|
-Supramolecule #1: AQP7 dimer of tetramers with inhibitor
| Supramolecule | Name: AQP7 dimer of tetramers with inhibitor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: sample was prepared in GDN detergent |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Aquaporin-7
| Macromolecule | Name: Aquaporin-7 / type: protein_or_peptide / ID: 1 / Details: LIG represents the inhibitor / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.264395 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MVQASGHRRS TRGSKMVSWS VIAKIQEILQ RKMVREFLAE FMSTYVMMVF GLGSVAHMVL NKKYGSYLGV NLGFGFGVTM GVHVAGRIS GAHMNAAVTF ANCALGRVPW RKFPVYVLGQ FLGSFLAAAT IYSLFYTAIL HFSGGQLMVT GPVATAGIFA T YLPDHMTL ...String: MVQASGHRRS TRGSKMVSWS VIAKIQEILQ RKMVREFLAE FMSTYVMMVF GLGSVAHMVL NKKYGSYLGV NLGFGFGVTM GVHVAGRIS GAHMNAAVTF ANCALGRVPW RKFPVYVLGQ FLGSFLAAAT IYSLFYTAIL HFSGGQLMVT GPVATAGIFA T YLPDHMTL WRGFLNEAWL TGMLQLCLFA ITDQENNPAL PGTEALVIGI LVVIIGVSLG MNTGYAINPS RDLPPRIFTF IA GWGKQVF SNGENWWWVP VVAPLLGAYL GGIIYLVFIG STIPREPLKL EDSVAYEDHG ITVLPKMGSH EPTISPLTPV SVS PANRSS VHPAPPLHES MALEHF UniProtKB: Aquaporin-7 |
-Macromolecule #2: ethyl 4-[(4-pyrazol-1-ylphenyl)methylcarbamoylamino]benzoate
| Macromolecule | Name: ethyl 4-[(4-pyrazol-1-ylphenyl)methylcarbamoylamino]benzoate type: ligand / ID: 2 / Number of copies: 8 / Formula: T60 |
|---|---|
| Molecular weight | Theoretical: 364.398 Da |
| Chemical component information |  ChemComp-T60: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: 20mA for glow discharge | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK II / Details: 3s blot, 3s wait, 0s drain time, 0 blot force. | ||||||||||||
| Details | 25uM inhibitor was supplemented into protein solution before grid freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 9002 / Average exposure time: 2.16 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial model (PDB ID:8AMX) was fitted locally into map by Chimera. |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 108 |
| Output model |  PDB-8c9h: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)