[English] 日本語
 Yorodumi
Yorodumi- EMDB-16316: Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in apo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in apo state (Alpo4_comb dataset 3) | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Cys-loop receptor / pentameric ligand-gated ion channel / Alpo / nAChR / MEMBRANE PROTEIN | ||||||||||||||||||
| Biological species |  Alvinella pompejana (invertebrata) Alvinella pompejana (invertebrata) | ||||||||||||||||||
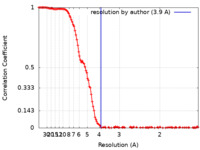
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||||||||
 Authors Authors | De Gieter S / Efremov RG / Ulens C | ||||||||||||||||||
| Funding support |  Belgium, 5 items Belgium, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Sterol derivative binding to the orthosteric site causes conformational changes in an invertebrate Cys-loop receptor. Authors: Steven De Gieter / Casey I Gallagher / Eveline Wijckmans / Diletta Pasini / Chris Ulens / Rouslan G Efremov /  Abstract: Cys-loop receptors or pentameric ligand-gated ion channels are mediators of electrochemical signaling throughout the animal kingdom. Because of their critical function in neurotransmission and high ...Cys-loop receptors or pentameric ligand-gated ion channels are mediators of electrochemical signaling throughout the animal kingdom. Because of their critical function in neurotransmission and high potential as drug targets, Cys-loop receptors from humans and closely related organisms have been thoroughly investigated, whereas molecular mechanisms of neurotransmission in invertebrates are less understood. When compared with vertebrates, the invertebrate genomes underwent a drastic expansion in the number of the nACh-like genes associated with receptors of unknown function. Understanding this diversity contributes to better insight into the evolution and possible functional divergence of these receptors. In this work, we studied orphan receptor Alpo4 from an extreme thermophile worm . Sequence analysis points towards its remote relation to characterized nACh receptors. We solved the cryo-EM structure of the lophotrochozoan nACh-like receptor in which a CHAPS molecule is tightly bound to the orthosteric site. We show that the binding of CHAPS leads to extending of the loop C at the orthosteric site and a quaternary twist between extracellular and transmembrane domains. Both the ligand binding site and the channel pore reveal unique features. These include a conserved Trp residue in loop B of the ligand binding site which is flipped into an apparent self-liganded state in the apo structure. The ion pore of Alpo4 is tightly constricted by a ring of methionines near the extracellular entryway of the channel pore. Our data provide a structural basis for a functional understanding of Alpo4 and hints towards new strategies for designing specific channel modulators. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16316.map.gz emd_16316.map.gz | 156.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16316-v30.xml emd-16316-v30.xml emd-16316.xml emd-16316.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16316_fsc.xml emd_16316_fsc.xml | 16.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_16316.png emd_16316.png | 80 KB | ||
| Masks |  emd_16316_msk_1.map emd_16316_msk_1.map | 166.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16316.cif.gz emd-16316.cif.gz | 6.3 KB | ||
| Others |  emd_16316_half_map_1.map.gz emd_16316_half_map_1.map.gz emd_16316_half_map_2.map.gz emd_16316_half_map_2.map.gz | 154.6 MB 154.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16316 http://ftp.pdbj.org/pub/emdb/structures/EMD-16316 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16316 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16316 | HTTPS FTP |
-Related structure data
| Related structure data |  8bxeMC  8bx5C  8bxbC  8bxdC  8bxfC  8byiC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16316.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16316.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7596 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16316_msk_1.map emd_16316_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16316_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16316_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pentameric complex of Alpo4 in apo state
| Entire | Name: Pentameric complex of Alpo4 in apo state |
|---|---|
| Components |
|
-Supramolecule #1: Pentameric complex of Alpo4 in apo state
| Supramolecule | Name: Pentameric complex of Alpo4 in apo state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Alvinella pompejana (invertebrata) Alvinella pompejana (invertebrata) |
| Molecular weight | Theoretical: 270 KDa |
-Macromolecule #1: Acetylcholine receptor
| Macromolecule | Name: Acetylcholine receptor / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Alvinella pompejana (invertebrata) Alvinella pompejana (invertebrata) |
| Molecular weight | Theoretical: 54.154934 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGSKVDKTL CRLGDITGLL IVLSVLVSRG LCQDCNNTNA SSMADEKRLL KCILHDYDTA IRPVQNVSDV VNVALEVTVV KVIDLDEKE HVLTTNGWIY HEWNDFQLKW NPSDYSGLKK IRIPVDRIWT PDIVLFNNAD ESYRYVVDKL AVVYYTGKVM W VPHARLRS ...String: MAGSKVDKTL CRLGDITGLL IVLSVLVSRG LCQDCNNTNA SSMADEKRLL KCILHDYDTA IRPVQNVSDV VNVALEVTVV KVIDLDEKE HVLTTNGWIY HEWNDFQLKW NPSDYSGLKK IRIPVDRIWT PDIVLFNNAD ESYRYVVDKL AVVYYTGKVM W VPHARLRS FCVLDLSRFP FDSQMCTLVF GSWTHDVSSV NVTLRNQSKV QYMIDGKEWQ VTSVQPKRYQ WTYNSNENYA GI ITGIKLK RTSIYYQYVF IMPTVLLAFL TLLMPFIPPL GKERITYGIG LVLGCTLLLM MLSDRMPTEL GNVPVVAAYL AYV FVMVAI NLLFAIMAIN MSMRDPKFGK VPGWVRWIFL TKLSKLVCLP VEPYTAVPAD LAYEETAAAR ELLDMNNGTA TADQ RVSGS DTKPALDRTL EDIRRYLRLV ATRTVVTPQL SHRDLVVQEW QQLTRVIDRL LFGSFLVLTV VITISMYAHY |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Phase plate: OTHER / Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 25669 / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)