[English] 日本語
 Yorodumi
Yorodumi- EMDB-16019: Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X0... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X04-labelled mice) | |||||||||
 Map data Map data | CryoEM map for extracted AppNLGF Abeta42 fibril after MOX04 labelling | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloid / fibril / helical / cross-beta / beta amyloid / PROTEIN FIBRIL / ex vivo / arctic mutant / alzheimers disease | |||||||||
| Function / homology |  Function and homology information Function and homology informationamyloid-beta complex / microglia development / negative regulation of presynapse assembly / cytosolic mRNA polyadenylation / collateral sprouting in absence of injury / synaptic assembly at neuromuscular junction / Formyl peptide receptors bind formyl peptides and many other ligands / regulation of Wnt signaling pathway / regulation of synapse structure or activity / axo-dendritic transport ...amyloid-beta complex / microglia development / negative regulation of presynapse assembly / cytosolic mRNA polyadenylation / collateral sprouting in absence of injury / synaptic assembly at neuromuscular junction / Formyl peptide receptors bind formyl peptides and many other ligands / regulation of Wnt signaling pathway / regulation of synapse structure or activity / axo-dendritic transport / axon midline choice point recognition / astrocyte activation involved in immune response / smooth endoplasmic reticulum calcium ion homeostasis / NMDA selective glutamate receptor signaling pathway / regulation of spontaneous synaptic transmission / mating behavior / ciliary rootlet / Golgi-associated vesicle / PTB domain binding / Lysosome Vesicle Biogenesis / positive regulation of amyloid fibril formation / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / neuron remodeling / Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models / nuclear envelope lumen / COPII-coated ER to Golgi transport vesicle / suckling behavior / positive regulation of protein metabolic process / dendrite development / modulation of excitatory postsynaptic potential / TRAF6 mediated NF-kB activation / presynaptic active zone / signaling receptor activator activity / Advanced glycosylation endproduct receptor signaling / The NLRP3 inflammasome / negative regulation of long-term synaptic potentiation / neuromuscular process controlling balance / transition metal ion binding / regulation of multicellular organism growth / intracellular copper ion homeostasis / regulation of presynapse assembly / negative regulation of neuron differentiation / ECM proteoglycans / spindle midzone / positive regulation of T cell migration / forebrain development / smooth endoplasmic reticulum / Purinergic signaling in leishmaniasis infection / positive regulation of chemokine production / Notch signaling pathway / clathrin-coated pit / positive regulation of G2/M transition of mitotic cell cycle / extracellular matrix organization / neuron projection maintenance / Mitochondrial protein degradation / axonogenesis / ionotropic glutamate receptor signaling pathway / positive regulation of mitotic cell cycle / response to interleukin-1 / positive regulation of calcium-mediated signaling / cholesterol metabolic process / protein serine/threonine kinase binding / platelet alpha granule lumen / positive regulation of glycolytic process / adult locomotory behavior / dendritic shaft / central nervous system development / positive regulation of long-term synaptic potentiation / positive regulation of interleukin-1 beta production / trans-Golgi network membrane / endosome lumen / learning / locomotory behavior / astrocyte activation / positive regulation of JNK cascade / Post-translational protein phosphorylation / microglial cell activation / serine-type endopeptidase inhibitor activity / synapse organization / regulation of long-term neuronal synaptic plasticity / positive regulation of non-canonical NF-kappaB signal transduction / TAK1-dependent IKK and NF-kappa-B activation / neuromuscular junction / visual learning / recycling endosome / positive regulation of interleukin-6 production / Golgi lumen / cognition / neuron cellular homeostasis / endocytosis / positive regulation of inflammatory response / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / cellular response to amyloid-beta / neuron projection development / G2/M transition of mitotic cell cycle / positive regulation of tumor necrosis factor production / apical part of cell / synaptic vesicle / Platelet degranulation / cell-cell junction Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Wilkinson M / Leistner C / Burgess A / Goodfellow S / Deuchars S / Ranson NA / Radford SE / Frank RAW | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The in-tissue molecular architecture of β-amyloid pathology in the mammalian brain. Authors: Conny Leistner / Martin Wilkinson / Ailidh Burgess / Megan Lovatt / Stanley Goodbody / Yong Xu / Susan Deuchars / Sheena E Radford / Neil A Ranson / René A W Frank /  Abstract: Amyloid plaques composed of Aβ fibrils are a hallmark of Alzheimer's disease (AD). However, the molecular architecture of amyloid plaques in the context of fresh mammalian brain tissue is unknown. ...Amyloid plaques composed of Aβ fibrils are a hallmark of Alzheimer's disease (AD). However, the molecular architecture of amyloid plaques in the context of fresh mammalian brain tissue is unknown. Here, using cryogenic correlated light and electron tomography we report the in situ molecular architecture of Aβ fibrils in the App familial AD mouse model containing the Arctic mutation and an atomic model of ex vivo purified Arctic Aβ fibrils. We show that in-tissue Aβ fibrils are arranged in a lattice or parallel bundles, and are interdigitated by subcellular compartments, extracellular vesicles, extracellular droplets and extracellular multilamellar bodies. The Arctic Aβ fibril differs significantly from an earlier App fibril structure, indicating a striking effect of the Arctic mutation. These structural data also revealed an ensemble of additional fibrillar species, including thin protofilament-like rods and branched fibrils. Together, these results provide a structural model for the dense network architecture that characterises β-amyloid plaque pathology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16019.map.gz emd_16019.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16019-v30.xml emd-16019-v30.xml emd-16019.xml emd-16019.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16019.png emd_16019.png | 91.5 KB | ||
| Filedesc metadata |  emd-16019.cif.gz emd-16019.cif.gz | 5.8 KB | ||
| Others |  emd_16019_half_map_1.map.gz emd_16019_half_map_1.map.gz emd_16019_half_map_2.map.gz emd_16019_half_map_2.map.gz | 56.6 MB 56.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16019 http://ftp.pdbj.org/pub/emdb/structures/EMD-16019 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16019 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16019 | HTTPS FTP |
-Validation report
| Summary document |  emd_16019_validation.pdf.gz emd_16019_validation.pdf.gz | 954.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16019_full_validation.pdf.gz emd_16019_full_validation.pdf.gz | 954 KB | Display | |
| Data in XML |  emd_16019_validation.xml.gz emd_16019_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_16019_validation.cif.gz emd_16019_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16019 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16019 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16019 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16019 | HTTPS FTP |
-Related structure data
| Related structure data |  8bfbMC  8bfaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16019.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16019.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map for extracted AppNLGF Abeta42 fibril after MOX04 labelling | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
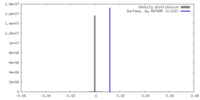
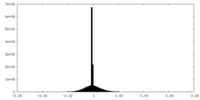
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: halfmap2
| File | emd_16019_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
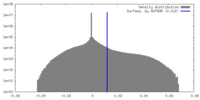
| Density Histograms |
-Half map: halfmap1
| File | emd_16019_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
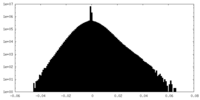
| Density Histograms |
- Sample components
Sample components
-Entire : Sarkosyl-extracted AppNL-G-F Abeta42 fibril
| Entire | Name: Sarkosyl-extracted AppNL-G-F Abeta42 fibril |
|---|---|
| Components |
|
-Supramolecule #1: Sarkosyl-extracted AppNL-G-F Abeta42 fibril
| Supramolecule | Name: Sarkosyl-extracted AppNL-G-F Abeta42 fibril / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Fibrils purified from mouse brain labelled with Methoxy-X04 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.441 kDa/nm |
-Macromolecule #1: Amyloid-beta precursor protein
| Macromolecule | Name: Amyloid-beta precursor protein / type: protein_or_peptide / ID: 1 Details: Humanised Abeta42 from App^NL-G-F mice with arctic mutation (E22G) Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.448025 KDa |
| Sequence | String: DAEFRHDSGY EVHHQKLVFF AGDVGSNKGA IIGLMVGGVV IA UniProtKB: Amyloid-beta precursor protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 6s blot. | |||||||||
| Details | Sarkosyl-insoluble fibrils from App^NL-G-F mouse brain labelled with Methoxy-X04 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 4165 / Average exposure time: 6.0 sec. / Average electron dose: 41.0 e/Å2 Details: 1442 raw EER frames were collected per image and combined into 40 fractions for processing |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 2.414 Å Applied symmetry - Helical parameters - Δ&Phi: 179.355 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 4.0) / Number images used: 3640 |
|---|---|
| Segment selection | Number selected: 136214 / Software - Name: crYOLO Details: Manually picked a subset of images to train a model for automatic fibril segment picking in crYOLO |
| Startup model | Type of model: INSILICO MODEL Details: Model generated from 2D class averages using relion_helix_inimodel2d |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: RELION (ver. 4.0) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 52 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-8bfb: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)