[English] 日本語
 Yorodumi
Yorodumi- EMDB-15984: Early transcription elongation state of influenza B/Mem polymeras... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Early transcription elongation state of influenza B/Mem polymerase backtracked due to double incoproation of nucleotide analogue T1106 and with singly incoporated T1106 at the U +1 position | |||||||||
 Map data Map data | LocScale map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza / viral RNA-dependent RNA polymerase / antiviral drug / nucleoside analogue / T705 (favipiravir) / T1106 / cap-dependent transcription / backtracking / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / virion component / endonuclease activity / host cell cytoplasm / Hydrolases; Acting on ester bonds / RNA-directed RNA polymerase ...cap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / virion component / endonuclease activity / host cell cytoplasm / Hydrolases; Acting on ester bonds / RNA-directed RNA polymerase / viral RNA genome replication / RNA-dependent RNA polymerase activity / nucleotide binding / DNA-templated transcription / host cell nucleus / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Influenza B virus (B/Memphis/13/2003) / Influenza B virus (B/Memphis/13/2003) /  Influenza B virus / Influenza B virus /  Influenza B virus (B/Acre/117700/2012) / Influenza B virus (B/Acre/117700/2012) /  Influenza B virus (B/Kaliningrad/RII06/2012) Influenza B virus (B/Kaliningrad/RII06/2012) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Cusack S / Kouba T | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: Direct observation of backtracking by influenza A and B polymerases upon consecutive incorporation of the nucleoside analog T1106. Authors: Tomas Kouba / Anna Dubankova / Petra Drncova / Elisa Donati / Pietro Vidossich / Valentina Speranzini / Alex Pflug / Johanna Huchting / Chris Meier / Marco De Vivo / Stephen Cusack /    Abstract: The antiviral pseudo-base T705 and its de-fluoro analog T1106 mimic adenine or guanine and can be competitively incorporated into nascent RNA by viral RNA-dependent RNA polymerases. Although ...The antiviral pseudo-base T705 and its de-fluoro analog T1106 mimic adenine or guanine and can be competitively incorporated into nascent RNA by viral RNA-dependent RNA polymerases. Although dispersed, single pseudo-base incorporation is mutagenic, consecutive incorporation causes polymerase stalling and chain termination. Using a template encoding single and then consecutive T1106 incorporation four nucleotides later, we obtained a cryogenic electron microscopy structure of stalled influenza A/H7N9 polymerase. This shows that the entire product-template duplex backtracks by 5 nt, bringing the singly incorporated T1106 to the +1 position, where it forms an unexpected T1106:U wobble base pair. Similar structures show that influenza B polymerase also backtracks after consecutive T1106 incorporation, regardless of whether prior single incorporation has occurred. These results give insight into the unusual mechanism of chain termination by pyrazinecarboxamide base analogs. Consecutive incorporation destabilizes the proximal end of the product-template duplex, promoting irreversible backtracking to a more energetically favorable overall configuration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15984.map.gz emd_15984.map.gz | 234.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15984-v30.xml emd-15984-v30.xml emd-15984.xml emd-15984.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15984_fsc.xml emd_15984_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15984.png emd_15984.png | 141.9 KB | ||
| Filedesc metadata |  emd-15984.cif.gz emd-15984.cif.gz | 7.7 KB | ||
| Others |  emd_15984_additional_1.map.gz emd_15984_additional_1.map.gz emd_15984_half_map_1.map.gz emd_15984_half_map_1.map.gz emd_15984_half_map_2.map.gz emd_15984_half_map_2.map.gz | 446.7 MB 382.4 MB 382.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15984 http://ftp.pdbj.org/pub/emdb/structures/EMD-15984 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15984 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15984 | HTTPS FTP |
-Validation report
| Summary document |  emd_15984_validation.pdf.gz emd_15984_validation.pdf.gz | 811.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15984_full_validation.pdf.gz emd_15984_full_validation.pdf.gz | 810.7 KB | Display | |
| Data in XML |  emd_15984_validation.xml.gz emd_15984_validation.xml.gz | 25.9 KB | Display | |
| Data in CIF |  emd_15984_validation.cif.gz emd_15984_validation.cif.gz | 34.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15984 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15984 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15984 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15984 | HTTPS FTP |
-Related structure data
| Related structure data |  8bdrMC  7qtlC  7r0eC  7r1fC  7zplC  7zpmC  8be0C  8bekC  8bf5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15984.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15984.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocScale map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.638 Å | ||||||||||||||||||||||||||||||||||||
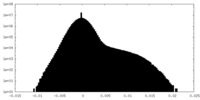
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Relion post-processed
| File | emd_15984_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion post-processed | ||||||||||||
| Projections & Slices |
| ||||||||||||
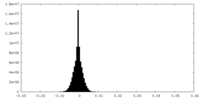
| Density Histograms |
-Half map: Half-map
| File | emd_15984_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
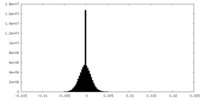
| Density Histograms |
-Half map: Half-map
| File | emd_15984_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
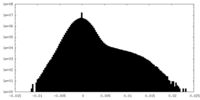
| Density Histograms |
- Sample components
Sample components
-Entire : Early transcription elongation state of influenza B/Mem polymeras...
| Entire | Name: Early transcription elongation state of influenza B/Mem polymerase backtracked due to double incoproation of nucleotide analogue T1106 and with singly incoporated T1106 at the U +1 position |
|---|---|
| Components |
|
-Supramolecule #1: Early transcription elongation state of influenza B/Mem polymeras...
| Supramolecule | Name: Early transcription elongation state of influenza B/Mem polymerase backtracked due to double incoproation of nucleotide analogue T1106 and with singly incoporated T1106 at the U +1 position type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Memphis/13/2003) Influenza B virus (B/Memphis/13/2003) |
-Macromolecule #1: Polymerase acidic protein
| Macromolecule | Name: Polymerase acidic protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Memphis/13/2003) / Strain: Influenza B virus (B/Memphis/13/2003) Influenza B virus (B/Memphis/13/2003) / Strain: Influenza B virus (B/Memphis/13/2003) |
| Molecular weight | Theoretical: 85.822781 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSHHHHHHHH GSGSMDTFIT RNFQTTIIQK AKNTMAEFSE DPELQPAMLF NICVHLEVCY VISDMNFLDE EGKAYTALEG QGKEQNLRP QYEVIEGMPR TIAWMVQRSL AQEHGIETPK YLADLFDYKT KRFIEVGITK GLADDYFWKK KEKLGNSMEL M IFSYNQDY ...String: GSHHHHHHHH GSGSMDTFIT RNFQTTIIQK AKNTMAEFSE DPELQPAMLF NICVHLEVCY VISDMNFLDE EGKAYTALEG QGKEQNLRP QYEVIEGMPR TIAWMVQRSL AQEHGIETPK YLADLFDYKT KRFIEVGITK GLADDYFWKK KEKLGNSMEL M IFSYNQDY SLSNESSLDE EGKGRVLSRL TELQAELSLK NLWQVLIGEE DVEKGIDFKL GQTISRLRDI SVPAGFSNFE GM RSYIDNI DPKGAIERNL ARMSPLVSVT PKKLTWEDLR PIGPHIYNHE LPEVPYNAFL LMSDELGLAN MTEGKSKKPK TLA KECLEK YSTLRDQTDP ILIMKSEKAN ENFLWKLWRD CVNTISNEEM SNELQKTNYA KWATGDGLTY QKIMKEVAID DETM CQEEP KIPNKCRVAA WVQTEMNLLS TLTSKRALDL PEIGPDVAPV EHVGSERRKY FVNEINYCKA STVMMKYVLF HTSLL NESN ASMGKYKVIP ITNRVVNEKG ESFDMLYGLA VKGQSHLRGD TDVVTVVTFE FSSTDPRVDS GKWPKYTVFR IGSLFV SGR EKSVYLYCRV NGTNKIQMKW GMEARRCLLQ SMQQMEAIVE QESSIQGYDM TKACFKGDRV NSPKTFSIGT QEGKLVK GS FGKALRVIFT KCLMHYVFGN AQLEGFSAES RRLLLLIQAL KDRKGPWVFD LEGMYSGIEE CISNNPWVIQ SAYWFNEW L GFEKEGSKVL ESVDEIMDEG SGSGENLYFQ UniProtKB: Polymerase acidic protein |
-Macromolecule #2: RNA-directed RNA polymerase catalytic subunit
| Macromolecule | Name: RNA-directed RNA polymerase catalytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Memphis/13/2003) / Strain: Influenza B virus (B/Memphis/13/2003) Influenza B virus (B/Memphis/13/2003) / Strain: Influenza B virus (B/Memphis/13/2003) |
| Molecular weight | Theoretical: 86.207016 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSGSGSGSGM NINPYFLFID VPIQAAISTT FPYTGVPPYS HGTGTGYTID TVIRTHEYSN KGKQYISDVT GCTMVDPTNG PLPEDNEPS AYAQLDCVLE ALDRMDEEHP GLFQAASQNA METLMVTTVD KLTQGRQTFD WTVCRNQPAA TALNTTITSF R LNDLNGAD ...String: GSGSGSGSGM NINPYFLFID VPIQAAISTT FPYTGVPPYS HGTGTGYTID TVIRTHEYSN KGKQYISDVT GCTMVDPTNG PLPEDNEPS AYAQLDCVLE ALDRMDEEHP GLFQAASQNA METLMVTTVD KLTQGRQTFD WTVCRNQPAA TALNTTITSF R LNDLNGAD KGGLIPFCQD IIDSLDRPEM TFFSVKNIKK KLPAKNRKGF LIKRIPMKVK DKITKVEYIK RALSLNTMTK DA ERGKLKR RAIATAGIQI RGFVLVVENL AKNICENLEQ SGLPVGGNEK KAKLSNAVAK MLSNCPPGGI SMTVTGDNTK WNE CLNPRI FLAMTERITR DSPIWFRDFC SIAPVLFSNK IARLGKGFMI TSKTKRLKAQ IPCPDLFSIP LERYNEETRA KLKK LKPFF NEEGTASLSP GMMMGMFNML STVLGVAALG IKNIGNKEYL WDGLQSSDDF ALFVNAKDEE TCMEGINDFY RTCKL LGIN MSKKKSYCNE TGMFEFTSMF YRDGFVSNFA MELPSFGVAG VNESADMAIG MTIIKNNMIN NGMGPATAQT AIQLFI ADY RYTYKCHRGD SKVEGKRMKI IKELWENTKG RDGLLVADGG PNIYNLRNLH IPEIVLKYNL MDPEYKGRLL HPQNPFV GH LSIEGIKEAD ITPAHGPVKK MDYDAVSGTH SWRTKRNRSI LNTDQRNMIL EEQCYAKCCN LFEACFNSAS YRKPVGQH S MLEAMAHRLR MDARLDYESG RMSKDDFEKA MAHLGEIGYI GSGSGENLYF Q UniProtKB: RNA-directed RNA polymerase catalytic subunit |
-Macromolecule #3: Polymerase basic protein 2
| Macromolecule | Name: Polymerase basic protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Memphis/13/2003) / Strain: Influenza B virus (B/Memphis/13/2003) Influenza B virus (B/Memphis/13/2003) / Strain: Influenza B virus (B/Memphis/13/2003) |
| Molecular weight | Theoretical: 90.844109 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSGSGSGSGM TLAKIELLKQ LLRDNEAKTV LKQTTVDQYN IIRKFNTSRI EKNPSLRMKW AMCSNFPLAL TKGDMANRIP LEYKGIQLK TNAEDIGTKG QMCSIAAVTW WNTYGPIGDT EGFERVYESF FLRKMRLDNA TWGRITFGPV ERVRKRVLLN P LTKEMPPD ...String: GSGSGSGSGM TLAKIELLKQ LLRDNEAKTV LKQTTVDQYN IIRKFNTSRI EKNPSLRMKW AMCSNFPLAL TKGDMANRIP LEYKGIQLK TNAEDIGTKG QMCSIAAVTW WNTYGPIGDT EGFERVYESF FLRKMRLDNA TWGRITFGPV ERVRKRVLLN P LTKEMPPD EASNVIMEIL FPKEAGIPRE STWIHRELIK EKREKLKGTM ITPIVLAYML ERELVARRRF LPVAGATSAE FI EMLHCLQ GENWRQIYHP GGNKLTESRS QSMIVACRKI IRRSIVASNP LELAVEIANK TVIDTEPLKS CLAAIDGGDV ACD IIRAAL GLKIRQRQRF GRLELKRISG RGFKNDEEIL IGNGTIQKIG IWDGEEEFHV RCGECRGILK KSKMKLEKLL INSA KKEDM RDLIILCMVF SQDTRMFQGV RGEINFLNRA GQLLSPMYQL QRYFLNRSND LFDQWGYEES PKASELHGIN ESMNA SDYT LKGVVVTRNV IDDFSSTETE KVSITKNLSL IKRTGEVIMG ANDVSELESQ AQLMITYDTP KMWEMGTTKE LVQNTY QWV LKNLVTLKAQ FLLGKEDMFQ WDAFEAFESI IPQKMAGQYS GFARAVLKQM RDQEVMKTDQ FIKLLPFCFS PPKLRSN GE PYQFLKLVLK GGGENFIEVR KGSPLFSYNP QTEVLTICGR MMSLKGKIED EERNRSMGNA VLAGFLVSGK YDPDLGDF K TIEELEKLKP GEKANILLYQ GKPVKVVKRK RYSALSNDIS QGIKRQRMTV ESMGWALSGW SHPQFEKGSG SENLYFQ UniProtKB: Polymerase basic protein 2 |
-Macromolecule #4: 5' vRNA
| Macromolecule | Name: 5' vRNA / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Influenza B virus Influenza B virus |
| Molecular weight | Theoretical: 4.49574 KDa |
| Sequence | String: AGUAGUAACA AGUU |
-Macromolecule #5: 3' vRNA
| Macromolecule | Name: 3' vRNA / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Acre/117700/2012) Influenza B virus (B/Acre/117700/2012) |
| Molecular weight | Theoretical: 6.699013 KDa |
| Sequence | String: UAUACAACUG AUGAGGCUAU U |
-Macromolecule #6: mRNA
| Macromolecule | Name: mRNA / type: rna / ID: 6 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Kaliningrad/RII06/2012) Influenza B virus (B/Kaliningrad/RII06/2012) |
| Molecular weight | Theoretical: 5.975564 KDa |
| Sequence | String: AUCUAUAAUA GCCUC(K1F)UC(K1F) |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #8: P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE
| Macromolecule | Name: P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE / type: ligand / ID: 8 / Number of copies: 1 / Formula: GTA |
|---|---|
| Molecular weight | Theoretical: 787.441 Da |
| Chemical component information | 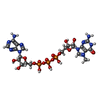 ChemComp-GTA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)