+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CRL4CSA-E2-Ub (state 2) | |||||||||
 Map data Map data | The main map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA repair / transcription / ubiqutin / cryo-EM | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of transcription-coupled nucleotide-excision repair / nucleotide-excision repair complex / negative regulation of granulocyte differentiation / response to auditory stimulus / single strand break repair / cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / (E3-independent) E2 ubiquitin-conjugating enzyme / cullin-RING ubiquitin ligase complex / Cul7-RING ubiquitin ligase complex ...regulation of transcription-coupled nucleotide-excision repair / nucleotide-excision repair complex / negative regulation of granulocyte differentiation / response to auditory stimulus / single strand break repair / cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / (E3-independent) E2 ubiquitin-conjugating enzyme / cullin-RING ubiquitin ligase complex / Cul7-RING ubiquitin ligase complex / regulation of DNA damage checkpoint / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / cellular response to chemical stress / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / double-strand break repair via classical nonhomologous end joining / regulation of nucleotide-excision repair / positive regulation of protein autoubiquitination / positive regulation by virus of viral protein levels in host cell / RNA polymerase II transcription initiation surveillance / protein neddylation / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / NEDD8 ligase activity / UV-damage excision repair / VCB complex / negative regulation of response to oxidative stress / Cul5-RING ubiquitin ligase complex / SCF ubiquitin ligase complex / ubiquitin-ubiquitin ligase activity / E2 ubiquitin-conjugating enzyme / negative regulation of type I interferon production / Cul2-RING ubiquitin ligase complex / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / Cul3-RING ubiquitin ligase complex / WD40-repeat domain binding / negative regulation of mitophagy / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Prolactin receptor signaling / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / ubiquitin conjugating enzyme activity / TGF-beta receptor signaling activates SMADs / negative regulation of reproductive process / negative regulation of developmental process / hemopoiesis / regulation of proteolysis / site of DNA damage / viral release from host cell / cullin family protein binding / somatic stem cell population maintenance / regulation of postsynapse assembly / anatomical structure morphogenesis / protein monoubiquitination / positive regulation of G1/S transition of mitotic cell cycle / response to X-ray / ectopic germ cell programmed cell death / positive regulation of viral genome replication / ubiquitin-like ligase-substrate adaptor activity / protein K48-linked ubiquitination / response to UV / protein autoubiquitination / proteasomal protein catabolic process / Nuclear events stimulated by ALK signaling in cancer / transcription-coupled nucleotide-excision repair / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / positive regulation of TORC1 signaling / regulation of cellular response to insulin stimulus / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / negative regulation of insulin receptor signaling pathway / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / positive regulation of gluconeogenesis / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / intrinsic apoptotic signaling pathway / post-translational protein modification / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / APC-Cdc20 mediated degradation of Nek2A / p75NTR recruits signalling complexes Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
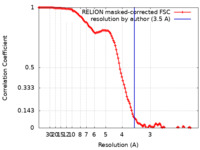
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Kokic G / Cramer P | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: C(N)RL4CSA-E2-Ub (state 2) Authors: Kokic G / Cramer P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15829.map.gz emd_15829.map.gz | 141.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15829-v30.xml emd-15829-v30.xml emd-15829.xml emd-15829.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15829_fsc.xml emd_15829_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_15829.png emd_15829.png | 129.7 KB | ||
| Filedesc metadata |  emd-15829.cif.gz emd-15829.cif.gz | 7.7 KB | ||
| Others |  emd_15829_half_map_1.map.gz emd_15829_half_map_1.map.gz emd_15829_half_map_2.map.gz emd_15829_half_map_2.map.gz | 139.3 MB 139.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15829 http://ftp.pdbj.org/pub/emdb/structures/EMD-15829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15829 | HTTPS FTP |
-Related structure data
| Related structure data |  8b3iMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15829.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15829.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The main map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
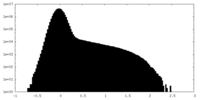
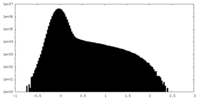
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map corresponding to the main map.
| File | emd_15829_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map corresponding to the main map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
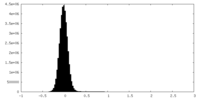
| Density Histograms |
-Half map: Half map corresponding to the main map.
| File | emd_15829_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map corresponding to the main map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
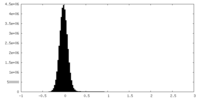
| Density Histograms |
- Sample components
Sample components
-Entire : C(N)RL4CSA-E2-Ub complex.
| Entire | Name: C(N)RL4CSA-E2-Ub complex. |
|---|---|
| Components |
|
-Supramolecule #1: C(N)RL4CSA-E2-Ub complex.
| Supramolecule | Name: C(N)RL4CSA-E2-Ub complex. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Ubiquitin-conjugating enzyme E2 D2
| Macromolecule | Name: Ubiquitin-conjugating enzyme E2 D2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: E2 ubiquitin-conjugating enzyme |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.755227 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALKRIHKEL NDLARDPPAQ CSAGPVGDDM FHWQATIMGP NDSPYQGGVF FLTIHFPTDY PFKPPKVAFT TRIYHPNINS NGSICLDIL RSQWSPALTI SKVLLSICSL LCDPNPDDPL VPEIARIYKT DREKYNRIAR EWTQKYAM UniProtKB: Ubiquitin-conjugating enzyme E2 D2 |
-Macromolecule #2: NEDD8
| Macromolecule | Name: NEDD8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.573978 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLIKVKTLTG KEIEIDIEPT DKVERIKERV EEKEGIPPQQ QRLIYSGKQM NDEKTAADYK ILGGSVLHLV LALRGG UniProtKB: Ubiquitin-like protein NEDD8 |
-Macromolecule #3: E3 ubiquitin-protein ligase RBX1
| Macromolecule | Name: E3 ubiquitin-protein ligase RBX1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.289977 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAAMDVDTP SGTNSGAGKK RFEVKKWNAV ALWAWDIVVD NCAICRNHIM DLCIECQANQ ASATSEECTV AWGVCNHAFH FHCISRWLK TRQVCPLDNR EWEFQKYGH UniProtKB: E3 ubiquitin-protein ligase RBX1 |
-Macromolecule #4: Ubiquitin
| Macromolecule | Name: Ubiquitin / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.576831 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN IQKESTLHLV LRLRGG UniProtKB: Polyubiquitin-C |
-Macromolecule #5: DNA excision repair protein ERCC-8
| Macromolecule | Name: DNA excision repair protein ERCC-8 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.10716 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MLGFLSARQT GLEDPLRLRR AESTRRVLGL ELNKDRDVER IHGGGINTLD IEPVEGRYML SGGSDGVIVL YDLENSSRQS YYTCKAVCS IGRDHPDVHR YSVETVQWYP HDTGMFTSSS FDKTLKVWDT NTLQTADVFN FEETVYSHHM SPVSTKHCLV A VGTRGPKV ...String: MLGFLSARQT GLEDPLRLRR AESTRRVLGL ELNKDRDVER IHGGGINTLD IEPVEGRYML SGGSDGVIVL YDLENSSRQS YYTCKAVCS IGRDHPDVHR YSVETVQWYP HDTGMFTSSS FDKTLKVWDT NTLQTADVFN FEETVYSHHM SPVSTKHCLV A VGTRGPKV QLCDLKSGSC SHILQGHRQE ILAVSWSPRY DYILATASAD SRVKLWDVRR ASGCLITLDQ HNGKKSQAVE SA NTAHNGK VNGLCFTSDG LHLLTVGTDN RMRLWNSSNG ENTLVNYGKV CNNSKKGLKF TVSCGCSSEF VFVPYGSTIA VYT VYSGEQ ITMLKGHYKT VDCCVFQSNF QELYSGSRDC NILAWVPSLY EPVPDDDETT TKSQLNPAFE DAWSSSDEEG UniProtKB: DNA excision repair protein ERCC-8 |
-Macromolecule #6: DNA damage-binding protein 1
| Macromolecule | Name: DNA damage-binding protein 1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.097469 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK ...String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK FLYGCQAPTI CFVYQDPQGR HVKTYEVSLR EKEFNKGPWK QENVEAEASM VIAVPEPFGG AIIIGQESIT YH NGDKYLA IAPPIIKQST IVCHNRVDPN GSRYLLGDME GRLFMLLLEK EEQMDGTVTL KDLRVELLGE TSIAECLTYL DNG VVFVGS RLGDSQLVKL NVDSNEQGSY VVAMETFTNL GPIVDMCVVD LERQGQGQLV TCSGAFKEGS LRIIRNGIGI HEHA SIDLP GIKGLWPLRS DPNRETDDTL VLSFVGQTRV LMLNGEEVEE TELMGFVDDQ QTFFCGNVAH QQLIQITSAS VRLVS QEPK ALVSEWKEPQ AKNISVASCN SSQVVVAVGR ALYYLQIHPQ ELRQISHTEM EHEVACLDIT PLGDSNGLSP LCAIGL WTD ISARILKLPS FELLHKEMLG GEIIPRSILM TTFESSHYLL CALGDGALFY FGLNIETGLL SDRKKVTLGT QPTVLRT FR SLSTTNVFAC SDRPTVIYSS NHKLVFSNVN LKEVNYMCPL NSDGYPDSLA LANNSTLTIG TIDEIQKLHI RTVPLYES P RKICYQEVSQ CFGVLSSRIE VQDTSGGTTA LRPSASTQAL SSSVSSSKLF SSSTAPHETS FGEEVEVHNL LIIDQHTFE VLHAHQFLQN EYALSLVSCK LGKDPNTYFI VGTAMVYPEE AEPKQGRIVV FQYSDGKLQT VAEKEVKGAV YSMVEFNGKL LASINSTVR LYEWTTEKEL RTECNHYNNI MALYLKTKGD FILVGDLMRS VLLLAYKPME GNFEEIARDF NPNWMSAVEI L DDDNFLGA ENAFNLFVCQ KDSAATTDEE RQHLQEVGLF HLGEFVNVFC HGSLVMQNLG ETSTPTQGSV LFGTVNGMIG LV TSLSESW YNLLLDMQNR LNKVIKSVGK IEHSFWRSFH TERKTEPATG FIDGDLIESF LDISRPKMQE VVANLQYDDG SGM KREATA DDLIKVVEEL TRIH UniProtKB: DNA damage-binding protein 1 |
-Macromolecule #7: Cullin-4A
| Macromolecule | Name: Cullin-4A / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 87.814297 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MADEAPRKGS FSALVGRTNG LTKPAALAAA PAKPGGAGGS KKLVIKNFRD RPRLPDNYTQ DTWRKLHEAV RAVQSSTSIR YNLEELYQA VENLCSHKVS PMLYKQLRQA CEDHVQAQIL PFREDSLDSV LFLKKINTCW QDHCRQMIMI RSIFLFLDRT Y VLQNSTLP ...String: MADEAPRKGS FSALVGRTNG LTKPAALAAA PAKPGGAGGS KKLVIKNFRD RPRLPDNYTQ DTWRKLHEAV RAVQSSTSIR YNLEELYQA VENLCSHKVS PMLYKQLRQA CEDHVQAQIL PFREDSLDSV LFLKKINTCW QDHCRQMIMI RSIFLFLDRT Y VLQNSTLP SIWDMGLELF RTHIISDKMV QSKTIDGILL LIERERSGEA VDRSLLRSLL GMLSDLQVYK DSFELKFLEE TN CLYAAEG QRLMQEREVP EYLNHVSKRL EEEGDRVITY LDHSTQKPLI ACVEKQLLGE HLTAILQKGL DHLLDENRVP DLA QMYQLF SRVRGGQQAL LQHWSEYIKT FGTAIVINPE KDKDMVQDLL DFKDKVDHVI EVCFQKNERF VNLMKESFET FINK RPNKP AELIAKHVDS KLRAGNKEAT DEELERTLDK IMILFRFIHG KDVFEAFYKK DLAKRLLVGK SASVDAEKSM LSKLK HECG AAFTSKLEGM FKDMELSKDI MVHFKQHMQN QSDSGPIDLT VNILTMGYWP TYTPMEVHLT PEMIKLQEVF KAFYLG KHS GRKLQWQTTL GHAVLKAEFK EGKKEFQVSL FQTLVLLMFN EGDGFSFEEI KMATGIEDSE LRRTLQSLAC GKARVLI KS PKGKEVEDGD KFIFNGEFKH KLFRIKINQI QMKETVEEQV STTERVFQDR QYQIDAAIVR IMKMRKTLGH NLLVSELY N QLKFPVKPGD LKKRIESLID RDYMERDKDN PNQYHYVA UniProtKB: Cullin-4A |
-Macromolecule #8: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 8 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.15 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)