[English] 日本語
 Yorodumi
Yorodumi- EMDB-15802: T5 Receptor Binding Protein pb5 in complex with its E. coli recep... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / receptor / complex / RPB / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsiderophore transmembrane transport / siderophore-iron import into cell / siderophore uptake transmembrane transporter activity / virus tail / transmembrane transporter complex / virion binding / toxic substance binding / cell outer membrane / signaling receptor activity / intracellular iron ion homeostasis ...siderophore transmembrane transport / siderophore-iron import into cell / siderophore uptake transmembrane transporter activity / virus tail / transmembrane transporter complex / virion binding / toxic substance binding / cell outer membrane / signaling receptor activity / intracellular iron ion homeostasis / entry receptor-mediated virion attachment to host cell / receptor-mediated virion attachment to host cell / iron ion binding / protein domain specific binding / symbiont entry into host cell / membrane Similarity search - Function | |||||||||
| Biological species |   Escherichia virus T5 / Escherichia virus T5 /  Escherichia phage T5 (virus) Escherichia phage T5 (virus) | |||||||||
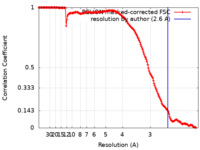
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Degroux S / Effantin G / Linares R / Schoehn G / Breyton C | |||||||||
| Funding support |  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2023 Journal: J Virol / Year: 2023Title: Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger. Authors: Séraphine Degroux / Grégory Effantin / Romain Linares / Guy Schoehn / Cécile Breyton /  Abstract: Bacteriophages, viruses infecting bacteria, recognize their host with high specificity, binding to either saccharide motifs or proteins of the cell wall of their host. In the majority of ...Bacteriophages, viruses infecting bacteria, recognize their host with high specificity, binding to either saccharide motifs or proteins of the cell wall of their host. In the majority of bacteriophages, this host recognition is performed by receptor binding proteins (RBPs) located at the extremity of a tail. Interaction between the RBPs and the host is the trigger for bacteriophage infection, but the molecular details of the mechanisms are unknown for most bacteriophages. Here, we present the electron cryomicroscopy (cryo-EM) structure of bacteriophage T5 RBP in complex with its Escherichia coli receptor, the iron ferrichrome transporter FhuA. Monomeric RBP is located at the extremity of T5's long flexible tail, and its irreversible binding to FhuA commits T5 to infection. Analysis of the structure of RBP within the complex, comparison with its AlphaFold2-predicted structure, and its fit into a previously determined map of the T5 tail tip in complex with FhuA allow us to propose a mechanism of transmission of the RBP receptor binding to the straight fiber, initiating the cascade of events that commits T5 to DNA ejection. Tailed bacteriophages specifically recognize their bacterial host by interaction of their receptor binding protein(s) (RBPs) with saccharides and/or proteins located at the surface of their prey. This crucial interaction commits the virus to infection, but the molecular details of this mechanism are unknown for the majority of bacteriophages. We determined the structure of bacteriophage T5 RBP in complex with its E. coli receptor, FhuA, by cryo-EM. This first structure of an RBP bound to its protein receptor allowed us to propose a mechanism of transmission of host recognition to the rest of the phage, ultimately opening the capsid and perforating the cell wall and, thus, allowing safe channeling of the DNA into the host cytoplasm. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15802.map.gz emd_15802.map.gz | 306.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15802-v30.xml emd-15802-v30.xml emd-15802.xml emd-15802.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15802_fsc.xml emd_15802_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15802.png emd_15802.png | 77 KB | ||
| Masks |  emd_15802_msk_1.map emd_15802_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15802.cif.gz emd-15802.cif.gz | 8.1 KB | ||
| Others |  emd_15802_half_map_1.map.gz emd_15802_half_map_1.map.gz emd_15802_half_map_2.map.gz emd_15802_half_map_2.map.gz | 277.6 MB 277.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15802 http://ftp.pdbj.org/pub/emdb/structures/EMD-15802 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15802 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15802 | HTTPS FTP |
-Related structure data
| Related structure data |  8b14MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15802.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15802.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.052 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15802_msk_1.map emd_15802_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
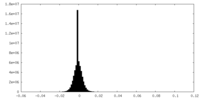
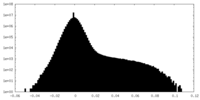
| Density Histograms |
-Half map: #2
| File | emd_15802_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15802_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between T5 Receptor Binding Protein pb5 and its E. coli r...
| Entire | Name: Complex between T5 Receptor Binding Protein pb5 and its E. coli receptor FhuA, stabilized with detergent |
|---|---|
| Components |
|
-Supramolecule #1: Complex between T5 Receptor Binding Protein pb5 and its E. coli r...
| Supramolecule | Name: Complex between T5 Receptor Binding Protein pb5 and its E. coli receptor FhuA, stabilized with detergent type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The FhuA-pb5 complex was formed by adding equimolar amounts of the two proteins, which results in 100% complex formation. FhuA-RBPpb5 complex is stabilized with 1.6% C10DAO at a protein ...Details: The FhuA-pb5 complex was formed by adding equimolar amounts of the two proteins, which results in 100% complex formation. FhuA-RBPpb5 complex is stabilized with 1.6% C10DAO at a protein concentration of 4.3 mg/mL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150 KDa |
-Supramolecule #2: E. coli receptor FhuA
| Supramolecule | Name: E. coli receptor FhuA / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Escherichia virus T5 Escherichia virus T5 |
-Supramolecule #3: T5 Receptor Binding Protein pb5
| Supramolecule | Name: T5 Receptor Binding Protein pb5 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: FhuA iron-ferrichrome transporter
| Macromolecule | Name: FhuA iron-ferrichrome transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 79.876945 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AVEPKEDTIT VTAAPAPQES AWGPAATIAA RQSATGTKTD TPIQKVPQSI SVVTAEEMAL HQPKSVKEAL SYTPGVSVGT RGASNTYDH LIIRGFAAEG QSQNNYLNGL KLQGNFYNDA VIDPYMLERA EIMRGPVSVL YGKSSPGGLL NMVSKRPTTE P LKEVQFKA ...String: AVEPKEDTIT VTAAPAPQES AWGPAATIAA RQSATGTKTD TPIQKVPQSI SVVTAEEMAL HQPKSVKEAL SYTPGVSVGT RGASNTYDH LIIRGFAAEG QSQNNYLNGL KLQGNFYNDA VIDPYMLERA EIMRGPVSVL YGKSSPGGLL NMVSKRPTTE P LKEVQFKA GTDSLFQTGF DFSDSLDDDG VYSYRLTGLA RSANAQQKGS EEQRYAIAPA FTWRPDDKTN FTFLSYFQNE PE TGYYGWL PKEGTVEPLP NGKRLPTDFN EGAKNNTYSR NEKMVGYSFD HEFNDTFTVR QNLRFAENKT SQNSVYGYGV CSD PANAYS KQCAALAPAD KGHYLARKYV VDDEKLQNFS VDTQLQSKFA TGDIDHTLLT GVDFMRMRND INAWFGYDDS VPLL NLYNP SHHHHHHGSV NTDFDFNAKD PANSGPYRIL NKQKQTGVYV QDQAQWDKVL VTLGGRYDWA DQESLNRVAG TTDKR DDKQ FTWRGGVNYL FDNGVTPYFS YSESFEPSSQ VGKDGNIFAP SKGKQYEVGV KYVPEDRPIV VTGAVYNLTK TNNLMA DPE GSFFSVEGGE IRARGVEIEA KAALSASVNV VGSYTYTDAE YTTDTTYKGN TPAQVPKHMA SLWADYTFFD GPLSGLT LG TGGRYTGSSY GDPANSFKVG SYTVVDALVR YDLARVGMAG SNVALHVNNL FDREYVASCF NTYGCFWGAE RQVVATAT F RF UniProtKB: Ferrichrome outer membrane transporter/phage receptor |
-Macromolecule #2: pb5 bacteriophage T5 receptor binding protein
| Macromolecule | Name: pb5 bacteriophage T5 receptor binding protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 68.782562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFFAGKLNN KSILSLRRGS GGDTNQHINP DSQTIFHSDM SHVIITETHS TGLRLDQGAG DYYWSEMPSR VTQLHNNDPN RVVLTEIEF SDGSRHMLSG MSMGVGAKAY GIINPQIMSQ GGLKTQITAS ADLSLDVGYF NTGTSGTIPQ KLRDGTGCQH M FGAFSGRR ...String: MSFFAGKLNN KSILSLRRGS GGDTNQHINP DSQTIFHSDM SHVIITETHS TGLRLDQGAG DYYWSEMPSR VTQLHNNDPN RVVLTEIEF SDGSRHMLSG MSMGVGAKAY GIINPQIMSQ GGLKTQITAS ADLSLDVGYF NTGTSGTIPQ KLRDGTGCQH M FGAFSGRR GFASSAMYLG GAALYKSAWS GSGYVVADAG TLTIPSDYVR HPGARNFGFN AIYVRGRSCN RVLYGMEGPN YT TGGAVQG ASSSGALNFT YNPSNPESPK YSVGFARADP TNYAYWESMG DPNDSANGPI GIYSEHLGIY PSKITWYVTN LVY NGSGYN IDGGLFNGND IKLSPREFII KGVNVNNTSW KFINFIEKNF NVGNRADFRD VGCNLSKDSP STGISGIATF GLPT TESNN APSIKGGNVG GLHANVVSIY NFLPSASWYV SSNPPKIGNN YGDVWSENLL PLRLLGGSGS TILSGNIVFQ GNGSV HVGT VGLDLNSSRN GAIVCTMEFI DDTWLSAGGI GCFNPTEMLS QGAEYGDSRF RIGGNTINKK LHQILSLPAG EYVPFF TIK GTVVNACKLQ AAAYNPTPYW VSGLPGSVGQ TGYYTLTYYM RNDGNNNISI WLDSSMSNII GMKACLPNIK LIIQRLT UniProtKB: Receptor-binding protein pb5 |
-Macromolecule #3: DECYLAMINE-N,N-DIMETHYL-N-OXIDE
| Macromolecule | Name: DECYLAMINE-N,N-DIMETHYL-N-OXIDE / type: ligand / ID: 3 / Number of copies: 1 / Formula: DDQ |
|---|---|
| Molecular weight | Theoretical: 201.349 Da |
| Chemical component information | 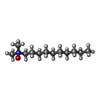 ChemComp-DDQ: |
-Macromolecule #4: [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethy...
| Macromolecule | Name: [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl] ...Name: [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate type: ligand / ID: 4 / Number of copies: 1 / Formula: LU9 |
|---|---|
| Molecular weight | Theoretical: 1.996235 KDa |
| Chemical component information |  ChemComp-LU9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.11111111111111 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| |||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER/RHODIUM / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: 25 mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV Details: 3.5 microliters of the FhuA-pb5 complex were deposited on a freshly glow discharged (25 mA, 30 sec) Cu/Rh 400 mesh Quantifoil R 2/1 EM grids and flash-frozen in nitrogen-cooled liquid ethane ...Details: 3.5 microliters of the FhuA-pb5 complex were deposited on a freshly glow discharged (25 mA, 30 sec) Cu/Rh 400 mesh Quantifoil R 2/1 EM grids and flash-frozen in nitrogen-cooled liquid ethane using a ThermoFisher Mark IV Vitrobot device (100% humidity, 293.15K, 2s blotting time, blot force 1).. | |||||||||
| Details | The FhuA-pb5 complex was formed by adding equimolar amounts of the two proteins, which results in 100% complex formation. FhuA-RBPpb5 complex is stabilized with 1.6% C10DAO at a protein concentration of 4.3 mg/ml |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Details | calibrated pixel size = 1.052 |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: COUNTING / #0 - Number grids imaged: 1 / #0 - Number real images: 8752 / #0 - Average electron dose: 60.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #1 - Detector mode: COUNTING / #1 - Number grids imaged: 1 / #1 - Number real images: 777 / #1 - Average electron dose: 60.0 e/Å2 #1 - Details: Unlike the first dataset, this one was acquired with a phase plate, close to focus (between -0.5 and -1.0 micrometer). |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The pb5 protein model was built de novo in the cryo-electron microscopy map. FhuA was adapted from the FhuA structure solved by X-ray crystallography (PDB 2GRX). The two structures were first refined separately using Coot (version 0.9.2) and Phenix (version 1.18.2-3874) softwares, then together. Structure validation was done using MolProbity. |
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8b14: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)