[English] 日本語
 Yorodumi
Yorodumi- EMDB-15713: Structure of Complement C5 in Complex with small molecule inhibit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Complement C5 in Complex with small molecule inhibitor and CVF | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Inhibitor / Protease / Complex / Cryo-EM / Toxin / IMMUNE SYSTEM | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationTerminal pathway of complement / membrane attack complex / Activation of C3 and C5 / negative regulation of macrophage chemotaxis / complement activation / complement activation, alternative pathway / chemokine activity / endopeptidase inhibitor activity / positive regulation of vascular endothelial growth factor production / complement activation, classical pathway ...Terminal pathway of complement / membrane attack complex / Activation of C3 and C5 / negative regulation of macrophage chemotaxis / complement activation / complement activation, alternative pathway / chemokine activity / endopeptidase inhibitor activity / positive regulation of vascular endothelial growth factor production / complement activation, classical pathway / positive regulation of chemokine production / Peptide ligand-binding receptors / Regulation of Complement cascade / chemotaxis / toxin activity / G alpha (i) signalling events / killing of cells of another organism / cell surface receptor signaling pathway / G protein-coupled receptor signaling pathway / inflammatory response / signaling receptor binding / extracellular space / extracellular exosome / extracellular region / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Naja kaouthia (monocled cobra) Naja kaouthia (monocled cobra) | ||||||||||||
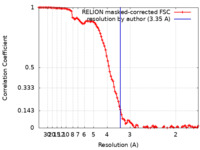
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | ||||||||||||
 Authors Authors | Srinivas H | ||||||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| ||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2019 Journal: Nat Chem Biol / Year: 2019Title: A small-molecule inhibitor of C5 complement protein. Authors: Keith Jendza / Mitsunori Kato / Michael Salcius / Honnappa Srinivas / Andrea De Erkenez / Anh Nguyen / Doug McLaughlin / Celine Be / Christian Wiesmann / Jason Murphy / Philippe Bolduc / ...Authors: Keith Jendza / Mitsunori Kato / Michael Salcius / Honnappa Srinivas / Andrea De Erkenez / Anh Nguyen / Doug McLaughlin / Celine Be / Christian Wiesmann / Jason Murphy / Philippe Bolduc / Muneto Mogi / Jose Duca / Abdel Namil / Michael Capparelli / Veronique Darsigny / Erik Meredith / Ritesh Tichkule / Luciana Ferrara / Jessica Heyder / Fang Liu / Patricia A Horton / Michael J Romanowski / Markus Schirle / Nello Mainolfi / Karen Anderson / Gregory A Michaud /   Abstract: The complement pathway is an important part of the immune system, and uncontrolled activation is implicated in many diseases. The human complement component 5 protein (C5) is a validated drug target ...The complement pathway is an important part of the immune system, and uncontrolled activation is implicated in many diseases. The human complement component 5 protein (C5) is a validated drug target within the complement pathway, as an anti-C5 antibody (Soliris) is an approved therapy for paroxysmal nocturnal hemoglobinuria. Here, we report the identification, optimization and mechanism of action for the first small-molecule inhibitor of C5 complement protein. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15713.map.gz emd_15713.map.gz | 8.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15713-v30.xml emd-15713-v30.xml emd-15713.xml emd-15713.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15713_fsc.xml emd_15713_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15713.png emd_15713.png | 51.4 KB | ||
| Filedesc metadata |  emd-15713.cif.gz emd-15713.cif.gz | 7.6 KB | ||
| Others |  emd_15713_half_map_1.map.gz emd_15713_half_map_1.map.gz emd_15713_half_map_2.map.gz emd_15713_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15713 http://ftp.pdbj.org/pub/emdb/structures/EMD-15713 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15713 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15713 | HTTPS FTP |
-Related structure data
| Related structure data |  8ayhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15713.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15713.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
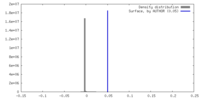
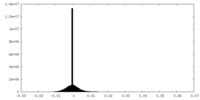
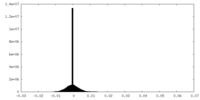
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15713_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
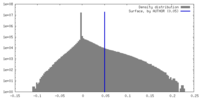
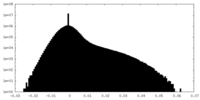
| Density Histograms |
-Half map: #2
| File | emd_15713_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
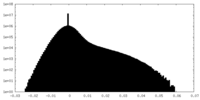
| Density Histograms |
- Sample components
Sample components
-Entire : Complement C5-Inhibitor-CVF complex
| Entire | Name: Complement C5-Inhibitor-CVF complex |
|---|---|
| Components |
|
-Supramolecule #1: Complement C5-Inhibitor-CVF complex
| Supramolecule | Name: Complement C5-Inhibitor-CVF complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 320 KDa |
-Macromolecule #1: Complement C5 beta chain
| Macromolecule | Name: Complement C5 beta chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.615766 KDa |
| Sequence | String: QEQTYVISAP KIFRVGASEN IVIQVYGYTE AFDATISIKS YPDKKFSYSS GHVHLSSENK FQNSAILTIQ PKQLPGGQNP VSYVYLEVV SKHFSKSKRM PITYDNGFLF IHTDKPVYTP DQSVKVRVYS LNDDLKPAKR ETVLTFIDPE GSEVDMVEEI D HIGIISFP ...String: QEQTYVISAP KIFRVGASEN IVIQVYGYTE AFDATISIKS YPDKKFSYSS GHVHLSSENK FQNSAILTIQ PKQLPGGQNP VSYVYLEVV SKHFSKSKRM PITYDNGFLF IHTDKPVYTP DQSVKVRVYS LNDDLKPAKR ETVLTFIDPE GSEVDMVEEI D HIGIISFP DFKIPSNPRY GMWTIKAKYK EDFSTTGTAY FEVKEYVLPH FSVSIEPEYN FIGYKNFKNF EITIKARYFY NK VVTEADV YITFGIREDL KDDQKEMMQT AMQNTMLING IAQVTFDSET AVKELSYYSL EDLNNKYLYI AVTVIESTGG FSE EAEIPG IKYVLSPYKL NLVATPLFLK PGIPYPIKVQ VKDSLDQLVG GVPVTLNAQT IDVNQETSDL DPSKSVTRVD DGVA SFVLN LPSGVTVLEF NVKTDAPDLP EENQAREGYR AIAYSSLSQS YLYIDWTDNH KALLVGEHLN IIVTPKSPYI DKITH YNYL ILSKGKIIHF GTREKFSDAS YQSINIPVTQ NMVPSSRLLV YYIVTGEQTA ELVSDSVWLN IEEKCGNQLQ VHLSPD ADA YSPGQTVSLN MATGMDSWVA LAAVDSAVYG VQRGAKKPLE RVFQFLEKSD LGCGAGGGLN NANVFHLAGL TFLTNAN AD DSQENDEPCK EILRP UniProtKB: Complement C5 |
-Macromolecule #2: Complement C5 alpha chain
| Macromolecule | Name: Complement C5 alpha chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 112.635008 KDa |
| Sequence | String: TLQKKIEEIA AKYKHSVVKK CCYDGACVNN DETCEQRAAR ISLGPRCIKA FTECCVVASQ LRANISHKDM QLGRLHMKTL LPVSKPEIR SYFPESWLWE VHLVPRRKQL QFALPDSLTT WEIQGVGISN TGICVADTVK AKVFKDVFLE MNIPYSVVRG E QIQLKGTV ...String: TLQKKIEEIA AKYKHSVVKK CCYDGACVNN DETCEQRAAR ISLGPRCIKA FTECCVVASQ LRANISHKDM QLGRLHMKTL LPVSKPEIR SYFPESWLWE VHLVPRRKQL QFALPDSLTT WEIQGVGISN TGICVADTVK AKVFKDVFLE MNIPYSVVRG E QIQLKGTV YNYRTSGMQF CVKMSAVEGI CTSESPVIDH QGTKSSKCVR QKVEGSSSHL VTFTVLPLEI GLHNINFSLE TW FGKEILV KTLRVVPEGV KRESYSGVTL DPRGIYGTIS RRKEFPYRIP LDLVPKTEIK RILSVKGLLV GEILSAVLSQ EGI NILTHL PKGSAEAELM SVVPVFYVFH YLETGNHWNI FHSDPLIEKQ KLKKKLKEGM LSIMSYRNAD YSYSVWKGGS ASTW LTAFA LRVLGQVNKY VEQNQNSICN SLLWLVENYQ LDNGSFKENS QYQPIKLQGT LPVEARENSL YLTAFTVIGI RKAFD ICPL VKIDTALIKA DNFLLENTLP AQSTFTLAIS AYALSLGDKT HPQFRSIVSA LKREALVKGN PPIYRFWKDN LQHKDS SVP NTGTARMVET TAYALLTSLN LKDINYVNPV IKWLSEEQRY GGGFYSTQDT INAIEGLTEY SLLVKQLRLS MDIDVSY KH KGALHNYKMT DKNFLGRPVE VLLNDDLIVS TGFGSGLATV HVTTVVHKTS TSEEVCSFYL KIDTQDIEAS HYRGYGNS D YKRIVACASY KPSREESSSG SSHAVMDISL PTGISANEED LKALVEGVDQ LFTDYQIKDG HVILQLNSIP SSDFLCVRF RIFELFEVGF LSPATFTVYE YHRPDKQCTM FYSTSNIKIQ KVCEGAACKC VEADCGQMQE ELDLTISAET RKQTACKPEI AYAYKVSIT SITVENVFVK YKATLLDIYK TGEAVAEKDS EITFIKKVTC TNAELVKGRQ YLIMGKEALQ IKYNFSFRYI Y PLDSLTWI EYWPRDTTCS SCQAFLANLD EFAEDIFLNG C UniProtKB: Complement C5 |
-Macromolecule #3: Cobra venom factor
| Macromolecule | Name: Cobra venom factor / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Naja kaouthia (monocled cobra) Naja kaouthia (monocled cobra) |
| Molecular weight | Theoretical: 184.726734 KDa |
| Sequence | String: MERMALYLVA ALLIGFPGSS HGALYTLITP AVLRTDTEEQ ILVEAHGDST PKQLDIFVHD FPRKQKTLFQ TRVDMNPAGG MLVTPTIEI PAKEVSTDSR QNQYVVVQVT GPQVRLEKVV LLSYQSSFLF IQTDKGIYTP GSPVLYRVFS MDHNTSKMNK T VIVEFQTP ...String: MERMALYLVA ALLIGFPGSS HGALYTLITP AVLRTDTEEQ ILVEAHGDST PKQLDIFVHD FPRKQKTLFQ TRVDMNPAGG MLVTPTIEI PAKEVSTDSR QNQYVVVQVT GPQVRLEKVV LLSYQSSFLF IQTDKGIYTP GSPVLYRVFS MDHNTSKMNK T VIVEFQTP EGILVSSNSV DLNFFWPYNL PDLVSLGTWR IVAKYEHSPE NYTAYFDVRK YVLPSFEVRL QPSEKFFYID GN ENFHVSI TARYLYGEEV EGVAFVLFGV KIDDAKKSIP DSLTRIPIID GDGKATLKRD TFRSRFPNLN ELVGHTLYAS VTV MTESGS DMVVTEQSGI HIVASPYQIH FTKTPKYFKP GMPYELTVYV TNPDGSPAAH VPVVSEAFHS MGTTLSDGTA KLIL NIPLN AQSLPITVRT NHGDLPRERQ ATKSMTAIAY QTQGGSGNYL HVAITSTEIK PGDNLPVNFN VKGNANSLKQ IKYFT YLIL NKGKIFKVGR QPRRDGQNLV TMNLHITPDL IPSFRFVAYY QVGNNEIVAD SVWVDVKDTC MGTLVVKGDN LIQMPG AAM KIKLEGDPGA RVGLVAVDKA VYVLNDKYKI SQAKIWDTIE KSDFGCTAGS GQNNLGVFED AGLALTTSTN LNTKQRS AA KCPQPANRRR RSSVLLLDSN ASKAAEFQDQ DLRKCCEDVM HENPMGYTCE KRAKYIQEGD ACKAAFLECC RYIKGVRD E NQRESELFLA RDDNEDGFIA DSDIISRSDF PKSWLWLTKD LTEEPNSQGI SSKTMSFYLR DSITTWVVLA VSFTPTKGI CVAEPYEIRV MKVFFIDLQM PYSVVKNEQV EIRAILHNYV NEDIYVRVEL LYNPAFCSAS TKGQRYRQQF PIKALSSRAV PFVIVPLEQ GLHDVEIKAS VQEALWSDGV RKKLKVVPEG VQKSIVTIVK LDPRAKGVGG TQLEVIKARK LDDRVPDTEI E TKIIIQGD PVAQIIENSI DGSKLNHLII TPSGCGEQNM IRMAAPVIAT YYLDTTEQWE TLGINRRTEA VNQIVTGYAQ QM VYKKADH SYAAFTNRAS SSWLTAYVVK VFAMAAKMVA GISHEIICGG VRWLILNRQQ PDGAFKENAP VLSGTMQGGI QGA EEEVYL TAFILVALLE SKTICNDYVN SLDSSIKKAT NYLLKKYEKL QRPYTTALTA YALAAADQLN DDRVLMAAST GRDH WEEYN AHTHNIEGTS YALLALLKMK KFDQTGPIVR WLTDQNFYGE TYGQTQATVM AFQALAEYEI QMPTHKDLNL DITIE LPDR EVPIRYRINY ENALLARTVE TKLNQDITVT ASGDGKATMT ILTFYNAQLQ EKANVCNKFH LNVSVENIHL NAMGAK GAL MLKICTRYLG EVDSTMTIID ISMLTGFLPD AEDLTRLSKG VDRYISRYEV DNNMAQKVAV IIYLNKVSHS EDECLHF KI LKHFEVGFIQ PGSVKVYSYY NLDEKCTKFY HPDKGTGLLN KICIGNVCRC AGETCSSLNH QERIDVPLQI EKACETNV D YVYKTKLLRI EEQDGNDIYV MDVLEVIKQG TDENPRAKTH QYISQRKCQE ALNLKVNDDY LIWGSRSDLL PTKDKISYI ITKNTWIERW PHEDECQEEE FQKLCDDFAQ FSYTLTEFGC PT UniProtKB: Cobra venom factor |
-Macromolecule #4: 5-methoxy-2-[[(1~{S})-1-(2-methoxyphenyl)ethyl]carbamoylamino]-4-...
| Macromolecule | Name: 5-methoxy-2-[[(1~{S})-1-(2-methoxyphenyl)ethyl]carbamoylamino]-4-(4-methylpentoxy)benzoic acid type: ligand / ID: 4 / Number of copies: 1 / Formula: H1H |
|---|---|
| Molecular weight | Theoretical: 444.521 Da |
| Chemical component information | 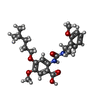 ChemComp-H1H: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50mM HEPES pH 7.5 100mM NaCl |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | Complement C5-Inhibitor-CVF complex |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 1.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)