+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pentameric ligand-gated ion channel GLIC with bound lipids | |||||||||
 Map data Map data | Refined map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GLIC / ion channel / pentameric channel / proton-gated channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsodium channel activity / potassium channel activity / extracellular ligand-gated monoatomic ion channel activity / transmembrane signaling receptor activity / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Gloeobacter violaceus (bacteria) Gloeobacter violaceus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Bergh C / Rovsnik U / Howard RJ / Lindahl E | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Discovery of lipid binding sites in a ligand-gated ion channel by integrating simulations and cryo-EM. Authors: Cathrine Bergh / Urška Rovšnik / Rebecca Howard / Erik Lindahl /  Abstract: Ligand-gated ion channels transduce electrochemical signals in neurons and other excitable cells. Aside from canonical ligands, phospholipids are thought to bind specifically to the transmembrane ...Ligand-gated ion channels transduce electrochemical signals in neurons and other excitable cells. Aside from canonical ligands, phospholipids are thought to bind specifically to the transmembrane domain of several ion channels. However, structural details of such lipid contacts remain elusive, partly due to limited resolution of these regions in experimental structures. Here, we discovered multiple lipid interactions in the channel GLIC by integrating cryo-electron microscopy and large-scale molecular simulations. We identified 25 bound lipids in the GLIC closed state, a conformation where none, to our knowledge, were previously known. Three lipids were associated with each subunit in the inner leaflet, including a buried interaction disrupted in mutant simulations. In the outer leaflet, two intrasubunit sites were evident in both closed and open states, while a putative intersubunit site was preferred in open-state simulations. This work offers molecular details of GLIC-lipid contacts particularly in the ill-characterized closed state, testable hypotheses for state-dependent binding, and a multidisciplinary strategy for modeling protein-lipid interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15649.map.gz emd_15649.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15649-v30.xml emd-15649-v30.xml emd-15649.xml emd-15649.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15649_fsc.xml emd_15649_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_15649.png emd_15649.png | 35.4 KB | ||
| Masks |  emd_15649_msk_1.map emd_15649_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15649.cif.gz emd-15649.cif.gz | 6.4 KB | ||
| Others |  emd_15649_additional_1.map.gz emd_15649_additional_1.map.gz emd_15649_additional_2.map.gz emd_15649_additional_2.map.gz emd_15649_half_map_1.map.gz emd_15649_half_map_1.map.gz emd_15649_half_map_2.map.gz emd_15649_half_map_2.map.gz | 7 MB 57.7 MB 49.8 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15649 http://ftp.pdbj.org/pub/emdb/structures/EMD-15649 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15649 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15649 | HTTPS FTP |
-Related structure data
| Related structure data |  8atgMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15649.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15649.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refined map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
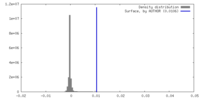
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15649_msk_1.map emd_15649_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
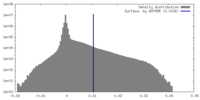
| Density Histograms |
-Additional map: Relion Postprocessed Map
| File | emd_15649_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion Postprocessed Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
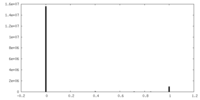
| Density Histograms |
-Additional map: Phenix autosharpen Map used for lipid building
| File | emd_15649_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phenix autosharpen Map used for lipid building | ||||||||||||
| Projections & Slices |
| ||||||||||||
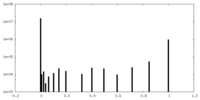
| Density Histograms |
-Half map: Half Map 2
| File | emd_15649_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1
| File | emd_15649_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pentameric ligand-gated ion channel
| Entire | Name: Pentameric ligand-gated ion channel |
|---|---|
| Components |
|
-Supramolecule #1: Pentameric ligand-gated ion channel
| Supramolecule | Name: Pentameric ligand-gated ion channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Gloeobacter violaceus (bacteria) Gloeobacter violaceus (bacteria) |
-Macromolecule #1: Proton-gated ion channel
| Macromolecule | Name: Proton-gated ion channel / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Gloeobacter violaceus (bacteria) / Strain: ATCC 29082 / PCC 7421 Gloeobacter violaceus (bacteria) / Strain: ATCC 29082 / PCC 7421 |
| Molecular weight | Theoretical: 36.29175 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AQDMVSPPPP IADEPLTVNT GIYLIECYSL DDKAETFKVN AFLSLSWKDR RLAFDPVRSG VRVKTYEPEA IWIPEIRFVN VENARDADV VDISVSPDGT VQYLERFSAR VLSPLDFRRY PFDSQTLHIY LIVRSVDTRN IVLAVDLEKV GKNDDVFLTG W DIESFTAV ...String: AQDMVSPPPP IADEPLTVNT GIYLIECYSL DDKAETFKVN AFLSLSWKDR RLAFDPVRSG VRVKTYEPEA IWIPEIRFVN VENARDADV VDISVSPDGT VQYLERFSAR VLSPLDFRRY PFDSQTLHIY LIVRSVDTRN IVLAVDLEKV GKNDDVFLTG W DIESFTAV VKPANFALED RLESKLDYQL RISRQYFSYI PNIILPMLFI LFISWTAFWS TSYEANVTLV VSTLIAHIAF NI LVETNLP KTPYMTYTGA IIFMIYLFYF VAVIEVTVQH YLKVESQPAR AASITRASRI AFPVVFLLAN IILAFLFFGF UniProtKB: Proton-gated ion channel |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 25 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 25 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.6 µm / Nominal defocus min: 2.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)